Translate this page into:
MicroRNA in carcinogenesis & cancer diagnostics: A new paradigm
Reprint requests: Dr Seyed E. Hasnain, School of Biological Sciences, Indian Institute of Technology, Delhi, Hauz Khas, New Delhi 110 016, India e-mail: seyedhasnain@gmail.com
-
Received: ,
This is an open-access article distributed under the terms of the Creative Commons Attribution-Noncommercial-Share Alike 3.0 Unported, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
This article was originally published by Medknow Publications & Media Pvt Ltd and was migrated to Scientific Scholar after the change of Publisher.
Abstract
MicroRNAs (miRNAs) are small 22-25 nucleotides long non-coding RNAs, that are conserved during evolution, and control gene expression in metazoan animals, plants, viruses, and bacteria primarily at post-transcriptional and transcriptional levels. MiRNAs ultimately regulate target gene expression by degrading the corresponding mRNA and/or inhibiting their translation. Currently, the critical functions of miRNAs have been established in regulating immune system, cell proliferation, differentiation and development, cancer and cell cycle by as yet unknown control mechanism. MiRNAs play an essential role in malignancy, and as tumour suppressors and oncogenes. Thus, discovery of new miRNAs will probably change the landscape of cancer genetics. Significantly different miRNA profiles can be assigned to various types of tumours, which could serve as phenotypic signatures for different cancers for their exploitation in cancer diagnostics, prognostics and therapeutics. If miRNA profiles can accurately predict malignancies, this technology could be exploited as a tool to surmount the diagnostic challenges. This review provides comprehensive and systematic information on miRNA biogenesis and their implications in human health.
Keywords
Biogenesis
cancer-gene expression
MicroRNA
oncogene
Introduction
MicroRNA (miRNA) was first discovered in 1993, and was characterized as a small, non-coding RNA (lin-4) molecule in Caenorhabditis elegans, regulating lin-14 protein expression12. About a few years later the second miRNA, let-7, was discovered in C. elegans3. These two discoveries of miRNA encouraged further studies on discovery of new miRNAs, and consequently a large class of small non-coding RNAs emerged with a diverse range of biological functions, such as temporal regulation of development, cell death and proliferation, hematopoiesis and tumourigenesis45. In general, metazoan miRNA genes are found within introns or exons, whereas most plant miRNAs are present within the intergenic region. Their complementary structure and length are also significantly variable. Animal miRNA genes are clustered within the genome and co-transcribed as polycistronic RNAs6. Thousands of miRNAs have been found in animals, plants, viruses, and an individual miRNA can post-transcriptionally target hundreds of mRNAs78. Till date more than 700 human miRNAs have been cloned. It has also been estimated that more than 45,000 miRNA target sites are present within human 3′ UTRs, and that more than 60 per cent of human protein-coding genes are possibly regulated by miRNAs. In the animal kingdom, about 30 per cent of all genes are targeted by miRNA910. Mature miRNA is ~22 nt short RNA molecule which is produced from a ~70 nt hairpin-like structure, called the precursor miRNA (pre-miRNA) that plays an important role in transcriptional regulation. It has been suggested that the miRNA genes could have more rapid turnover rate than protein-coding genes. However, each miRNA is thought to regulate multiple genes and hundreds of target genes have been predicted. Most miRNAs verified with important functions are highly conserved among species. However, adaptive evolution of lineage specific miRNAs has also been identified in diverse species11.
MiRNAs discovery led to a worldwide research effort to establish their roles in cancer. MiRNAs regulate molecular pathways in cancer by targeting various oncogenes and tumour suppressors, and have a role in cancer and stem cell biology, angiogenesis, the epithelial–mesenchymal transition, metastasis, and drug resistance. The let-7 miRNA family has a role in cancer by negatively regulating let-60/RAS12. Administration of miR-26a using adeno-associated virus (AAV) in an animal model of hepatocellular carcinoma (HCC) inhibits tumour progression without toxicity13. Anti-miRNA oligonucleotides (AMOs), can block the interactions between miRNA and its target mRNAs through competitive inhibition of base-pairing14. Administration of AMOs against miR-16, miR-122, miR-192 and miR-194 in animals offer efficient and sustained silencing of corresponding miRNAs15. There is currently no cost-effective screening test for Non-small cell lung cancer (NSCLC). NSCLC could be differentiated from healthy controls using a combination of differentially expressed miRNAs miR-15b and miR-27b, thereby pointing to the potentials of serum miRNAs as biomarkers for early detection of NSCLC16. Anti-miR-21 inhibited growth and migration of MCF-7 and MDA-MB-231 cells in vitro, and tumour growth in nude mice. Knockdown of miR-21 significantly increased the expression of ANKRD46 at both mRNA and protein levels revealed that miR-21 directly targeted ANKRD46. miR-21 and EIF4A2 protein are inversely expressed in breast cancers17. MiR-214 plays important roles in the pathogenesis of cervical cancer. It binds to the 3’UTR of UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 7 (GALNT7), thereby repressing GALNT7 expression18. Many cellular processes including cell proliferation, migration, invasion and survival have been directly linked to overexpression of miR-2119, whereas, its down regulation induces apoptosis20. Overexpression of miR-155, another commonly dysregulated miRNA, is also directly associated with tumourigenesis in lymphomas, breast, lung, colon, and pancreatic cancers21.
The problems of stability, specificity and delivery of short oligonucleotides are serious impediments in clinical applications of miRNA-based therapeutics. However, with the development of LNA (locked nucleic acid) technology, a phase I clinical trial with LNA-anti-miR-122, based on exciting preliminary data on non-human primates, has raised considerable hopes of a breakthrough21. The delivery and testing of miRNA mimics, using cationic liposomes or polymer-based nanoparticle formulations, have raised hopes of a paradigm shift in medicine and pharmaceutical industry. However, limited in vivo studies have been reported till date in this specific area22. An improved understanding of miRNA mechanisms in tumourigenesis and cancer maintenance would thus provide invaluable information about key cancer pathways, cancer diagnostics, and disease prognosis. This knowledge could be used in the development of anticancer therapies.
The significance of miRNAs and the small RNA (sRNAs) has been recognized across the living kingdom, including their roles in regulating immune response and host-pathogen interactions during bacterial, fungal and viral infections, and response to environmental stress have been reviewed elsewhere232425. This review is focused on miRNA biogenesis, their specific targets and functions in human cancer.
Biogenesis of miRNA
Most of the miRNA genes, similar to class II genes, are transcribed by RNA polymerase II (Pol II) to generate a stem loop primary miRNA (pri-miRNA), which can vary in size from a few hundred nucleotides to several kilobases. However, some miRNAs are also transcribed by RNA polymerase III, found in repetitive elements2627. About 40 per cent of miRNA loci are present in the intronic region and about 10 per cent in the exonic region of non-coding transcripts, and approximately 40 per cent are localized to the introns of protein-coding genes while the remaining miRNA genes are located in other regions5. Alternative splicing determines whether a miRNA is intronic or exonic. MiRNAs are 5’capped and 3’ polyadenylated and may also be spliced similar to mRNAs28. Mainly mammalian miRNAs, encoded in introns suggests that these are spliced before miRNA processing6. The pri-miRNAs are processed within the nucleus by a number of proteins called the ‘microprocessors’, of which RNase III enzyme, Drosha, double-stranded RNA binding (dsRBD) protein and cofactor DGCR8/Pasha are the core components (Fig. 1)2930. This complex cleaves the pri-miRNA stem by guiding the distance from the single-stranded/double-stranded RNA junction producing a 70-nt hairpin precursor miRNA called (pre-miRNA). The 2-nt 3’overhang at the cleavage site, characteristic of RNase III–mediated cleavage, is recognized by Exportin-5, a nuclear export factor, in a Ran-GTP dependent manner, which transports the pre-miRNA into the cytoplasm3132. Within the cytoplasm, the pre-miRNA is cleaved to produce the mature22-nt miRNA:miRNA duplex by another RNase III enzyme (Dicer), which interacts with the dsRBD, human immunodeficiency virus (HIV) transactivating response RNA binding protein (TRBP) and protein activator of the interferon induced protein kinase (PACT)3334. In human cells, TRBP binds the Argonaute protein (Ago2) and instantaneously with Dicer to form a trimeric complex. This initiates the assembly of the RNA-induced silencing complex (RISC), a ribonucleoprotein complex that leads to mRNA degradation3536. The miRNA strand with lower stability of base-pairing at its 5’end is incorporated into RISC complex, and another strand is typically degraded. Deep-sequencing revealed that the average ratio of miRNA to miRNA*as 100:1, and can be much lower in cases when both strands are functional37.
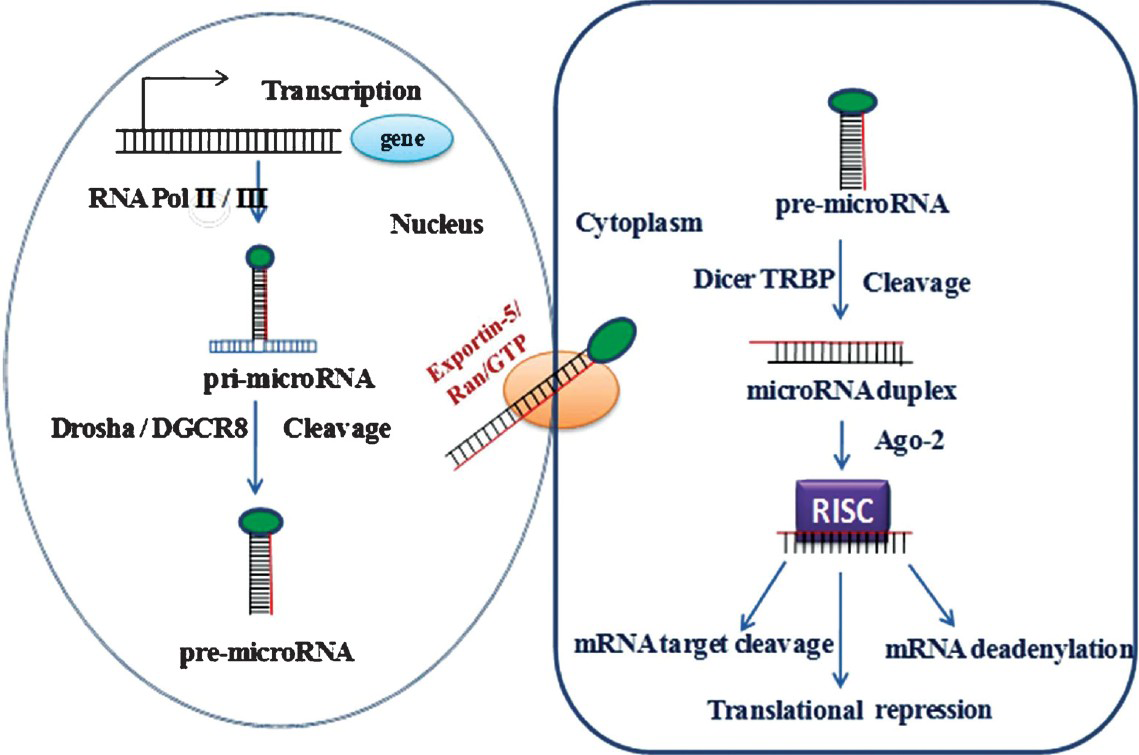
- MicroRNA pathway is universal to all mammalian miRNAs. MiRNAs are generated through two step processing that converts primary miRNA transcript (pri-miRNA) by RNA polymerase II or III and cleavage of the pri-miRNA by the microprocessor complex Drosha– DGCR8 (Pasha) in the nucleus. The resulting miRNA is exported from the nucleus by Exportin-5-Ran-GTP. In the cytoplasm the RNase Dicer, in complex with the double-stranded RNA-binding protein TRBP, cleaves the pre-miRNA hairpin to its mature length. Association of the miRNA-RISC with a target mRNA results in the repression of the target gene by promoting mRNA degradation and/or translational inhibition.
Once incorporated into RISC, the miRNA guides the complex to target the RNA by base-pairing interactions. In case of animals, the binding is perfect or near-perfect complementarily to the miRNA, this target mRNA is cleaved and degraded, otherwise, translation is repressed38. RISC contains an Ago2 protein capable of endonucleolytic cleavage. Ago2 is the sole enzyme conferring this activity in mammals and is the major enzyme in flies39. Most animal miRNAs base-pairing is imperfect with their targets, which stimulates translational repression rather than cleavage and degradation. In this type of repression, target mRNAs are not actively degraded; however, these can be destabilized by deadenylation and subsequent decapping840.
The mechanism of translational repression by miRNAs is not clearly understood. The step at which miRNAs block translation is controversial. There is evidence that miRNAs blocks translation initiation, whereas other studies suggest a block in elongation4142. Ago2 proteins bound to miRNAs and their target mRNAs accumulate in processing bodies (P-bodies), which represent cytoplasmic foci that are known sites of mRNA degradation43. The P-bodies exclude ribosomal components and are the sites in which mRNAs can be stored without translation. Numerous proteins found in P-bodies are GW182, Dcp1/Dcp2 decapping complex, and RCK/p54 helicase that can bind to Argonaute protein, and this interaction mediates translational repression40434445. While most attention has been on miRNA action in the cytoplasm, a report shows that mature miR-29b contains a 6-nt motif at its 3’ terminus, which directs the import of mature miRNA into the nucleus46, raising intriguing possibilities for other modes of miRNA function.
Interaction of MiRNAs with target gene
The criteria for miRNA target interactions were defined by mutation of known miRNA-target sites and testing for function in miRNA misexpression assays94748. These studies are focused on the importance of pairing of the 5’ end of the miRNA to mRNA target site, called the seed region48.
Computational studies followed by experimental validation have highlighted the importance of seed region in miRNA-target recognition. Such studies showed significant conservation of matches to miRNA seeds or, in some cases, absence of miRNA seed matches49. Single-nucleotide polymorphism (SNP) genotype data revealed that polymorphism density was significantly lower in conserved target site regions that are complementary to the 5’ region of the miRNA50. On the basis of miRNA-target prediction rules derived from experimental approaches and based on evolutionary conserved sequence, it has been estimated that more than 30 per cent of animal genes may be targeted by miRNA51. It has been predicted that the miRNA-target sites, without relying on cross-species conservation or miRNA sequence, consist of a larger number of miRNA-regulated genes52.
Target predictions have also been made by miRNA misexpression and target downregulation. Overexpression of miRNAs in tissue culture followed by expression profiling provides a global picture of target RNAs that are destabilized by miRNA binding7. However, many potential targets are not affected at the RNA level, suggesting that these approaches will underestimate meaningful miRNA-target relationships. At the protein level, miRNA target interactions are not amenable to high throughput approaches and must be tested one by one in reporter assays. Although miRNA misexpression is useful for testing possible regulation, yet it is not sufficient to draw conclusions about miRNA-target relationships in vivo. Given the large numbers of predicted miRNA-target genes and the lack of genetic evidence, miRNA-target relationship remains an open question. The SNPs computational studies revealed that within the conserved miRNA sites, 85 per cent sites are functionally important50.
Animal miRNAs could downregulate their target genes by translational repression, cleavage, or destabilization. Generally, it was believed that the animal miRNAs mostly function by way of translational repression whereas the plant miRNAs function via post-transcriptional gene silencing53545556. This hypothesis is supported by the differences in the levels of complementarity between the miRNA and its target in animals and plants. In plants, miRNAs are normally complimentary to the entire target site on the mRNA of the gene that these regulate54. In animals, however, only about seven nucleotides of the miRNA, known as seed region, complementary to the target site are sufficient to trigger miRNA-mRNA interaction. It is found that an individual miRNA (miR-1 or miR-124) could downregulate expression of hundreds of target genes directly when it is overexpressed in HeLa cells7. It has been argued that animal miRNAs can directly lead to target mRNA downregulation by mRNA cleavage55, and accelerated deadenylation of mRNAs8. It has been suggested that individual miRNA could directly repress the protein levels of hundreds of genes56. Animal miRNAs have also been found to induce translation upregulation of target mRNAs upon cell cycle arrest or bind the 5’UTR of ribosomal protein mRNAs and enhance their translation57. Guo et al58 suggested the miRNA mediated regulation gene expression through the destabilization of target mRNA, which was considered as a predominant reason for reduced translation.
Computational analysis data predict that miRNA targets with much ease in plants than in animals. This is due to the fact that miRNAs complementarity between targeted mRNAs is much higher in plants than in animals for a majority of their targets sequences59. Thus, in plants majority of miRNAs targets have been predicted and some of these have also been validated experimentally. However, such approaches have not been successful in identifying targets for many of the animal miRNAs. There are only one or a few targets for a majority of plant miRNAs54.
Overexpression and misexpression of miRNA
Transfection of the miRNAs (miR-1 and miR-124) into HeLa cells and subsequent expression profiling identified 100-200 affected transcripts7. Interestingly, the transcripts targeted were those that would normally be expressed at a low level, in which the miRNA is expressed. For instance, RNAs targeted by the muscle-specific miR-1, upon their transfection into non muscle cells were normally expressed at undetectable levels in muscle. Global analyses of miRNA expression patterns to those of their conserved targets arrived at a similar conclusion60. In Drosophila many miRNAs and their targets appear to be expressed in a largely non-overlapping manner, either temporally or spatially. In case of the latter, the targets are typically present in domains adjacent to the miRNA-expressing tissues. The mammalian cells expression profiling studies have shown that many conserved targets are indeed present in tissues expressing the miRNA60.
Despite the abundance of miRNA genes, a few miRNA mutants have been recovered using a multitude of genetic screens in Drosophila or C. elegans. In most cases, the seven nucleotides that constitute the seed region must be affected to eliminate miRNA function, making miRNAs as a difficult to hit targets in chemical mutagenesis61. Another probability is that many miRNA mutants show defects or low-penetrance defects that may be difficult to identify in high-throughput genetic screenings61.
Misexpressed miRNA in a cell shuts down endogenous expression of all of its target genes causing strong phenotype change60. However, miRNA and the target gene may not be co-expressed at the same time, rendering extrapolation of results of effect on the normal function of the miRNA rather difficult. miRNA misexpression can produce interesting defects that have limited application to what could be seen from miRNA mutant studies62.
The Enhancer of split-Complex and the Bearded-Complex genes, which are two Drosophila Notch target gene families, have conserved sequences in their 3’UTRs that are complementary to the seed sequence of related miRNA family. Misexpression of some of these miRNAs result in phenotypes similar to that of loss of Notch function. Misexpression of miR-iab-5p can repress Ubx and induce a homeotic phenotype63. However, it remains to be determined whether the corresponding miRNAs mutants will impact Notch signaling or Ubx function in vivo. While the misexpression results seem to have limited predictive value, in some cases, it may help to identify the correct target gene62.
Overexpression of the pancreatic islet-specific miR-375 results in inhibition of glucose-induced insulin secretion. This can be mimicked by knockdown of its target, myotrophin gene64. MiR-375 inversely regulates myotrophin expression and enhances glucose-stimulated insulin secretion. Using cultured hippocampal neuronic cells miR-134 was found to regulate dendritic spine size by inhibiting translation of Lim k165. Induced expression of B cell-specific miRNA, miR-181, in hematopoietic stem cells results in their differentiation to B-lineage cells, however the effect of loss of miR-181 on B-cell differentiation is not known. Overexpression and depletion of miR-181 produces opposing effects on antigen sensitivity in T cells66.
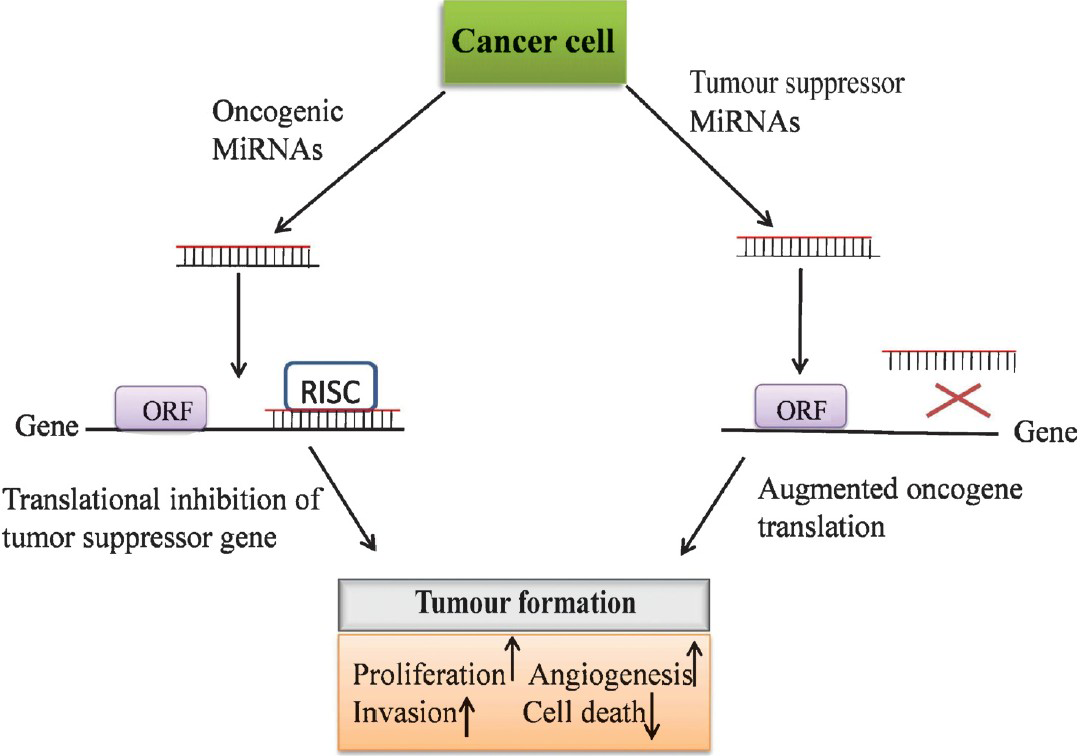
- MicroRNAs as oncogenes or tumour suppressor genes.
MiR-1 and miR-133 are absent from undifferentiated myoblasts and strongly upregulated upon differentiation into myotubes67. Tissue specific overexpression of miR-1 in the developing mouse heart induces premature differentiation of myocytes68. MiR-1 misexpression can accelerate myoblast differentiation by targeting histone deacetylase 4, a repressor of muscle differentiation, whereas depletion of miR-1impedes differentiation, as evident from a decrease in myogenic markers50. The miR-1 and miR-133 form one genomic cluster are coexpressed in heart and skeletal muscle. MiR-133 promotes myoblast proliferation by targeting serum response factor50, but in a different experimental setup myoblast differentiation was not observed67. During differentiation, miR-133 downregulated nPTB protein, a repressor of alternative splicing, causing an aberrant splicing of a group of silenced exons in mature myotubes. Both miR-1 and miR-133 are apparently required to maintain the properties of differentiated muscle cells7. The importance of miRNA family in cardiogenesis69 was evident from studies where deletion in one of the miR-1 genes resulted in defects in heart development and function.
In C. elegans, let-60/RAS contains several putative miR-84/let-7-binding sites and can be downregulated by miR-84and let-7. let-7 mutants display a burst vulva phenotype that can be suppressed by RNAi of let-60/RAS12, suggesting that excess of let-60/RAS activity contributes to the defect. Over-expression of miR-84, a member of the let-7 family, can rescue the multivulva phenotype caused by let-60/RAS gain-of-function alleles12. While these results indicate that let-7 could be an important in vivo regulator of let-60/RAS, definitive conclusions about the relevance of miR-84 await mutant analysis.
MiRNAs diagnosis as oncogene and tumour suppressor
Development of cancer entails combined interaction of both tumour suppressors and cancer inducers. miRNAs may function as a novel class of oncogenes and tumour suppressor genes70. The miRNAs with increased expression in tumours are thought to function as oncogenes and are termed as oncomirs. These oncomirs negatively inhibit tumour suppressor genes and/or those controlling cell differentiation or apoptosis and thereby promote tumour development. In contrast, some miRNAs exhibit decreased expression in cancerous cells and are considered as tumour suppressor genes. Tumuor suppressor miRNAs usually prevent tumour development by negatively inhibiting oncogenes and/or genes that control cell differentiation or apoptosis. miRNA let-7 is one of the founding members of the miRNA family and is highly conserved70. let-7 is localized to a region of the chromosome, which is usually deleted in human cancers. The highest levels of let-7 expression occur in differentiated adult tissues and its inappropriate expression results in oncogenic loss of differentiation. The let-7 family of miRNAs is downregulated in many tumours, including lung and breast cancer71. Many of the let-7 family members are located in fragile genomic areas associated with lung, breast, and cervical cancer. Further, let-7 family members functionally inhibit the mRNAs of well characterized oncogenes, such as the Ras family74, HMGA275, and c-myc75 induced apoptosis and cell cycle arrest when overexpressed in lung and colon cancer in Burkitt lymphoma cell lines7375. Similarly, the miR-15a and miR-16-1 genes are located in chromosome region 13q14, which is deleted in most cases of chronic lymphocytic leukemia76. These miRNAs target the Bcl2, an anti-apoptotic gene, suggesting that loss of miR-15a and miR-16-1in B cells may lead to the inhibition of apoptosis, giving rise to malignancies77. Also, these miRNAs are overexpressed in pancreatic tumour. Ectopic expression of miR-16-1 negatively regulates cell growth and cell cycle progression and induces apoptosis in several human cancer cell lines77. Further, the miR-29 family members have been shown to be downregulated in chronic myeloid leukemia (CLL), lung cancer, invasive breast cancer, acute myeloid leukemia (AML), and cholangiocarcinoma6476777879. miR-34a has been reported to be downregulated in human glioma tumours and exhibited a role of potential tumour suppressor in brain tumours by targeting multiple oncogenic pathways and induced the differentiation of cancer stem cell80.
Despite the absence of direct experimental evidence or data from miRNA knockout mice many other miRNAs are considered to function as tumour suppressors. Interestingly, miRNAs with demonstrated role in tumour suppression, such as miR-15-a/16-1, miR-29s, and let-7, are present at more than one genomic locations and may indeed be differentially regulated. In HeLa cells, while the product of miR-29b-1/miR-29a locus on chromosome 7q32 is actively transcribed to generate the mature miR-29b, transcription from miR-29b-2/miR-29c locus on chromosome 1q23 is silenced46. The observed redundancy of miRNA genomic copy could be a strategy to provide evolutionary advantage by ensuring a function in situations where one allele is deleted or silenced. Even though these are derived from different precursors the mature miRNA product is identical.
MiR-155 was first described as tumour oncogene8182 and has been found to be highly expressed in paediatric Burkitt lymphoma, Hodgkin disease83, primary mediastinal non-Hodgkin lymphoma82, CLL83, AML80, lung cancer72, and breast cancer65. It is localized in a non-coding region within the B cell integration cluster (BIC) on chromosome 21q23, which cooperates with c-myc in oncogenesis. In in-vitro cultured chicken embryo fibroblasts when BIC and c-myc are co-expressed, using replication-competent retrovirus vectors, enhancement of growth is observed80.
Information on the mechanism of miR-155/BIC overexpression in cancer is very scanty. There is a positive correlation of miR-155 in AML, with high white counts together with FLT3 in tandem duplication (ITD) mutations. FLT3 inhibitor mediated blocking of FLT3-ITD signaling in human leukemic cells had no effect on miR-155 levels pointing to a FLT3-ITD independent expression of mir-15579. Using a transgenic mouse model overexpressing miR-155 in B cells, a polyclonal preleukemic pre–B cell proliferation was evident and this was followed by full blown B cell malignancy thereby demonstrating the role of miRNA in early leukemogenesis84. The critical role of miR-155 in defective dendritic cell functions, impaired cytokine secretion, and shifting the Th cell bias toward Th2 differentiation in mouse have been similarly demonstrated using knockout studies85.
Several lines of evidences demonstrating upregulation of miR-21 in a number of haematological malignancies such as AML79, CLL83, and solid tumours including glioblastoma, cancers of the liver, pancreas, prostate, stomach, colon, lung, breast648687, etc. convincingly argue in favour of its role as an oncogene. Anti-sense mediated silencing of miR-21 expression in cultured liver glioblastoma and breast cancer cells, causes an inhibition of cell growth and triggers activation of caspases resulting in increased apoptosis8788, by targeting tumour suppressor genes such as PTEN (phosphatase and tensin homolog)88, PDCD4 (programmed cell death 4)88, and TPM1(tropomyosin 1)89.
The miR-17-92 cluster, which is trans-activated by c-myc oncogene90, is located within 800 base pairs in the non-coding gene C13 or f25 at chromosome locus 13q31.3. This region has been reported to be frequently amplified in follicular lymphoma and diffuse large B cell lymphoma91. Members of the miR-17-92cluster, which comprises six miRNAs (miR-17, miR-18a, miR-19a, miR-20a, miR-19b-1, and miR-92-1), are highly expressed in a variety of solid tumours and haematological malignancies, including cancers of the breast, colon, lung, pancreas, prostate, and stomach as well as lymphomas92. These are also known to have important role in lung development93 and in regulation of the immune and hematopoietic systems94, and are involved in promoting proliferation, inhibiting apoptosis, inducing tumour angiogenesis, and cooperating with c-mycto caused lymphoma in mice9295. This cluster is essential for B cell proliferation and that a modest overexpression in B cells induces a lymphoproliferative disorder in mice has been convincingly demonstrated using gain and loss-of-function experiments94. The ability of this cluster to regulate cell cycle and cell proliferation is in part a reflection of its effect on E2F transcription factors, notably E2F1, E2F2, and E2F3, which activate multiple genes resulting in cell cycle progression from G1 to S phase96. Conversely, both E2F1 and E2F3 can activate the miR-17-92 cluster, establishing a regulatory loop90. This mir-17-92 cluster also downregulates anti-apoptotic proteins, Bim and the tumour suppressors PTEN and p29497. Similar to miR-155, this cluster also induces lymphoid proliferation causing a predisposition to secondary genetic abnormalities, eventually leading to a full-blown malignancy. Downregulation of miR-17-5p in breast cancer cell lines has been reported98, which when restored to normal expression caused a translational inhibition of the AIB1 (amplified in breast cancer) gene thereby resulting in decreased breast cancer proliferation. In 16.5 per cent of ovarian cancers, 21.9 per cent of breast cancers, and 20 per cent of melanomas miR-17-92 genomic locus was found to be deleted9299. A recent report has also highlighted the ability of miR-22, a miRNA induced by 12-O-tetradecanoylphorbol-12-acetate (TPA), to repress the Myc-Max transcriptional complex causing an accumulation of cells in the G1 phase of cell cycle99.
From the above observations, it appears that the expression of individual miRNAs in this cluster is regulated by a fine-tuned post-transcriptional mechanism. The data also support a tumour suppressor role for miR-17-5p, which seem to contradict earlier reports consistently showing upregulation of this cluster in cancer. This dual role (oncogene and tumour suppressor) has also been described in protein-coding genes involved in the pathogenesis of cancer, such as TP53100. Thus, it is possible that a miRNA depending on the tissue and its transcriptome, including the miRNA targets can act either as an oncogene or as a tumour suppressor. While screening for miRNAs that cooperate with oncogenes in cellular transformation, two miRNAs, miR-372 and miR-373 were identified. These miRs directly inhibited the expression of tumour suppressor gene LATS2 and induced proliferation and tumourigenesis in cooperation with Ras by neutralizing wild-type gene TP53101. This mechanism was also evident during the oncogenesis of human testicular germ cell tumors, where the wild-type TP53 pathway was targeted. The status of miRNAs showing the characteristics of both the oncogene and tumour suppressor is summarized in the Table.
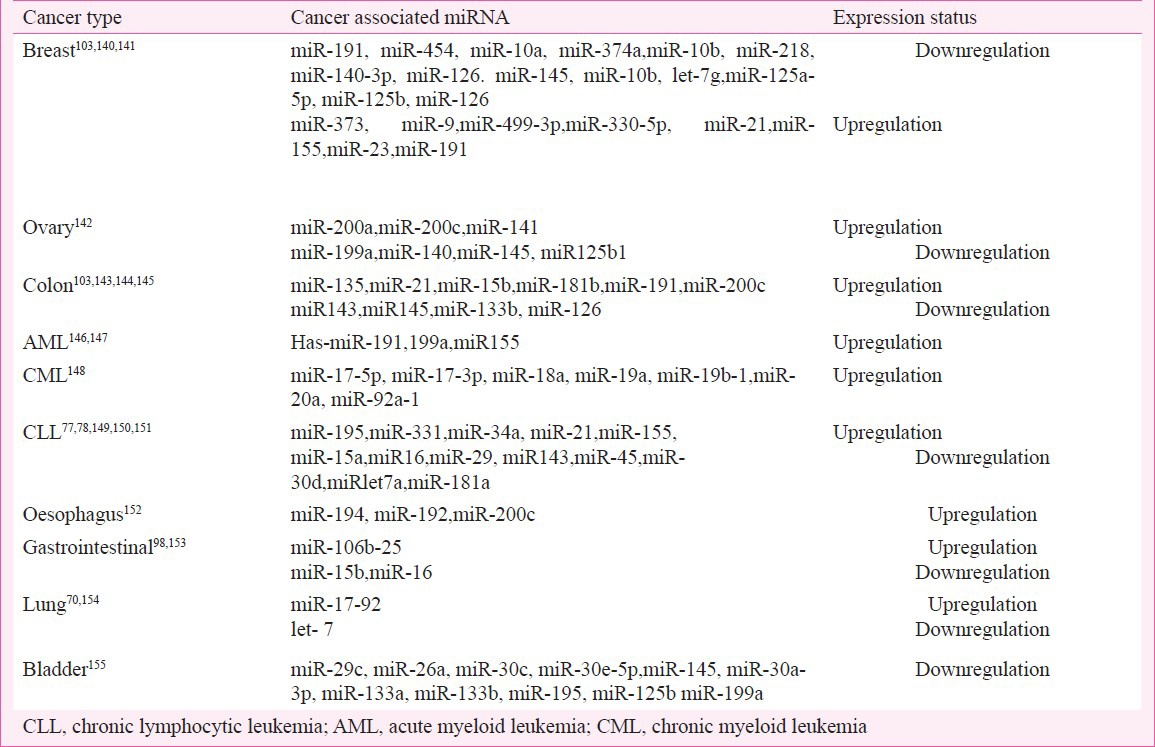
miRNA in cancer pathogenesis and diagnosis
The ability of miRNAs to regulate cell growth and apoptosis has found natural application in cancer pathogenesis which itself is a consequence of dysregulation of growth and apoptosis of cells. In order to identify the role of miRNAs in cancer pathogenesis, specific miRNAs overexpression or misexpresion can be studied to elucidate initiation and development of different types of malignancies. Recognition of miRNAs that are differentially expressed between tumour and normal tissues may help to identify those miRNAs that are involved in human cancers and further establish the role of miRNAs as biomarkers in cancer diagnostics102. For example, the reduced expression levels of let-7, observed both in vitro and in vivo studies; and its significant association with shortened post-operative survival, independent of disease stage point to the its role in lung cancer pathogenesis103. Similarly in breast cancer, expression levels of some miRNA such as miR-125b, miR-145, miR-21, and miR-155 were significantly reduced in neoplastic breast tissues as compared to normal tissues102. In breast cancer, dysregulation of miR-145 and miR-121 has been associated with tumour progression, whilst reduced let-7 expression has been associated with increased lymph node metastasis, suggesting its potential role as a diagnostic biomarker. The miRNAs miR-21 and miR-155 have also been suggested as biomarkers for diagnosis of non-small cell lung cancer104. Specific biopathologic features of breast cancers, such as tumour stage, proliferation index, estrogen and progesterone receptor expression, and vascular invasion have been correlated with miRNA expression levels. Colorectal neoplasia (colon cancer) is also associated with alteration in miRNA expression. About 28 different miRNAs have been identified in colonic adenocarcinoma and normal mucosa, and the expression of two mature miRNAs, miR-143, and miR-145, have been reported to be consistently reduced at the adenomatous and cancer stages of colorectal neoplasia105.
In primary human tumours, some reports have shown lower expression of miRNA than in normal tissues while others do not. Failure of Drosha processing has also been attributed to expression of many miRNA in differentiated cell types. Different types of tumours exhibit significantly different miRNA profiles pointing to their diagnostic and prognostic potential106107. While most miRNAs are not known to play an active role in tumurogenesis, their expression patterns have been increasingly associated with cancer initiation and progression and could serve as phenotypic signatures of different cancers thereby pointing to their diagnostic, prognostic and therapeutic value108109110111112113114.
MicroRNAs in cancer therapeutics
MiRNAs are known to be involved in several cellular processes such as apoptosis, proliferation, receptor-driven pathways, etc. MiRNAs delivery into target tissues, their stability and effectiveness into target tissues remain a major difficulty for direct RNA-based therapy. For cancer therapy introduction of miR-15a/16-1 induces apoptosis in leukaemic MEG01 cells and inhibits tumour growth115, while silencing oncogenic miR-21 with antisense oligonucleotides generates a proapoptotic and anti-proliferative response in vitro in different cellular models, reducing tumuor development and metastatic potential in vivo116. MiR-34a transiently inhibits human colon cancer progression when administered subcutaneously in complexes with atelocollagen, which was recently shown to be a very powerful system for efficient in vivo delivery of small interfering RNA molecules into tumours117. MiR-34, one of the best characterized tumour suppressor miRNAs known so far, functions downstream of p53 and upregulates genes involved in cell cycle arrest, cellular senescence and apoptosis. Given the association of defects in p53 pathway in a majority (>50%) of human cancers, miR-34 replacement therapy holds great clinical promise as a powerful therapeutic to treat patients with solid tumours118.
The susceptibility of unmodified dsRNAs to nucleases present in vivo is a major concern about their therapeutic use. Adenoviral vector mediated intranasal expression of let-7a RNA hairpin has been shown to cause a reduction of tumour formation in a lung cancer model, where conditional expression of oncogenic K-ras could be activated119. In agreement with their earlier reports that MYC induced liver tumours concomitantly downregulates a number of microRNAs120. Inhibition of cancer cell proliferation and induction of tumour-specific apoptosis using adeno-associated virus (AVV) vector mediated delivery of miR-26a in a mouse model of HCC119. Expression of foetal genes was also considerably reduced after overexpression of miR-133 and caused size reduction of left ventricular cardiac myocytes121. In another study, the initial observations on the crucial role of miR-10b as metastmiR in breast cancer was followed up by demonstrating its therapeutic potential in the form of suppression of breast cancer metastasis after treatment of tumour bearing mice systemically with miR-10b antagomirs122.
A placebo controlled, double-blind, randomized, single dose, dose-escalating safety study of SPC3649 involving 64 healthy male volunteers (LNA-antimiR-122; ClinicalTrials.gov, Identifier: NCT00688012) represents the first human clinical trial of LNA-anti-miR. Human liver is abundant with miR-122. Two closely spaced miR-122 target sites are present within the non-coding region, required for viral replication, in the Hepatitis C virus genome. A dose-dependent depletion of mature miR-122 was evident after delivery of LNAanti-miR in mice/and non-human primates123124.
Nanoparticles assisted miRNA delivery as a novel cancer therapeutic approach
The miRNAs and small interfering RNAs (siRNAs) are used to treat cancer by targeting specific protein by inhibiting their counterpart mRNA in mechanism of proliferation, invasion, antiapoptosis, drug resistance, and metastasis106125. Nanoparticles have been recently used to deliver the anti-miRNA for target genes suppression. Given the reported overexpression of miRNA-10b (mir-10b) in breast cancer cells and the associated cell migration and invasion, via inhibition of HOXD10 target synthesis, strategies to deliver the anti-miR-10b RNA molecule into breast cancer cells have clinical promise. A novel RNA poly L-lysine (PLL) complex has been developed which releases trace amounts of anti-mir-10b slowly for weeks into breast cancer cells with very high efficacy and without any toxicity126. LPH (liposome-polycation-hyaluronic acid) nanoparticles, modified with single-chain antibody fragment (scFv), for targeted systemic delivery of siRNA and miRNA into murine B16F10 melanoma, could also effectively downregulate target genes (c-Myc/MDM2/VEGF) in lung metastasis. The miRNA-34a (miR-34a) induced apoptosis is also associated with inhibition of survivin expression and downregulation of MAPK pathway in B16F10 cells127.
The delivery of RNA-based therapeutics for cancer therapy still remains a daunting task. Effective delivery of candidate genes to specific cell types is perhaps the biggest challenge in the field of gene therapy. Lately, the poly (D, L-lactide-co-glycolide) (PLGA)- based nanoparticles have been used for transfection of HepG2 cells with miRNA. This approach has overcome the disadvantages of cytotoxicity and aggregation on the cell surface, as were noticed with polyethylenimine (PEI) DNA complex. After transfection, PLGA nanocomplexes have been found to accumulate in tumour cells, and could enhance the effect of miR-26a by inducing cell cycle arrest128. miRNAs have been reported as cancer biomarkers that regulate tumour suppressor genes. Using a AS1411 aptamer and miRNA-221 molecular beacon (miR-221 MB)-conjugated magnetic fluorescence (MF) nanoparticle (MFAS miR-221 MB) was developed to simultaneously target cancer tissue129. Selective targeting and delivery of AS1411 aptamer-conjugated MF (MFAS) nanoparticles into various cancer cell lines were achieved. The miR-221 MB was detached from the MFAS miR-221 MB in the cytoplasm of C6 cells to reveal images of miRNA-221 biogenesis and simultaneously displayed its anti-tumour therapeutic potential. MFAS miRNA MB has also found application in other cancers by employing target miRNA highly expressed in the corresponding cancers113. Efficient delivery of anti-miRNA oligonucleotides (AMO) has also been demonstrated using functionalized gold nanoparticles (AuNPs) to silence miRNA miR-29b130. Delivery of AuNPs into HeLa cells to target miR-29b, significantly increased MCL-1 protein and mRNA levels and inhibited apoptosis induced by tumour necrosis factor-related apoptosis-inducing ligand (TRAIL). AuNPs can also deliver other AMOs against miRNA in other cell type130.
Differential miRNA expression may be consequences of epigenetic modification. The histone modification has been reported to be performed by cadmium telluride (CdTe) quantum dots (QDs) exposure131132. It suggests the possibility of involving miRNAs in the cytotoxicity of CdTe QDs. In addition, according to the non-genotoxic assessment, the apoptosis-like cell death is commonly induced by CdTe QDs in many cell lines133. The phenomenon suggests that miRNAs play a role in the cytotoxicity of CdTe QDs at the post-transcriptional level. Involvement of epigenetic mechanisms in miRNA biogenesis suggests that miRNAs act as participants in the cytotoxicity of CdTe QDs resulting in the apoptosis-like cell death134. Another therapeutic targets of cancer therapy involves PEGylated (covalent attachment of polyethylene glycol polymer chains to another molecule like drug or therapeutic protein) LPH (liposome-polycation-hyaluronic acid) nanoparticle formulations135. The nano-complex modified with cyclic RGD peptide (cRGD) has been used for specific and efficient endothelial cell delivery of anti-miRNA oligonucleotides (AMO) for targeting α(v)β integrin present on the tumour neovasculature. The delivered anti-miR-296 AMO to the cytoplasm reduced the expression of target miRNA in human umbilical vein endothelial cells (HUVECs). Consequently, hepatocyte growth factor-regulated tyrosine kinase substrate (HGS) was upregulated resulting in significantly suppressed blood tube formation and endothelial cell migration. On the other hand, nanoparticles lacking cRGD modification exhibited very little AMO uptake and miRNA silencing activity135.
Conclusions
miRNAs are now firmly established as potent post-transcriptional regulators of gene expression by simultaneously modulating a number of target genes. Ever since the first miRNA lin-4 was discovered in C. elegans and RNA interference (RNAi)136, there has been an exponential increase in our knowledge about the mechanism of miRNA-mediated gene regulation and their role in disease. Though we are still far away from getting a clear understanding of the precise roles of various miRNAs vis-à-vis their modulation of specific cellular processes, but it is clear that the biological roles of miRNA in development and disease, as well as their modes of action may vary in different biological conditions. What has convincingly emerged is the association of dysregulation of miRNAs with cancer initiation and progression, possibly through the modulation of apoptosis, and their likely role in cancer diagnosis, prognosis and therapy.
References
- The C. elegansheterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843-54.
- [Google Scholar]
- Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell. 1993;75:855-62.
- [Google Scholar]
- The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditiselegans. Nature. 2000;403:901-6.
- [Google Scholar]
- Micromanagers of gene expression: the potentially widespread influence of metazoan microRNAs. Nat Rev Genet. 2004;5:396-400.
- [Google Scholar]
- Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 2005;433:769-73.
- [Google Scholar]
- MicroRNAs direct rapid deadenylation of mRNA. Proc Natl Acad Sci USA. 2006;103:4034-9.
- [Google Scholar]
- Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15-20.
- [Google Scholar]
- Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19:92-05.
- [Google Scholar]
- MicroRNA regulation and the variability of human cortical gene expression. Nucleic Acids Res. 2008;36:4621-8.
- [Google Scholar]
- Therapeutic microRNA delivery suppresses tumorigenesis in a murine liver cancer model. Cell. 2009;137:1005-17.
- [Google Scholar]
- Anti-miRNA oligonucleotides (AMOs): ammunition to target miRNAs implicated in human disease? Gene Ther. 2006;13:496-502.
- [Google Scholar]
- Serum microRNA Biomarkers for Detection of Non-Small Cell Lung Cancer. PLoS ONE. 2012;7:e32307.
- [Google Scholar]
- Knockdown of miR-21 in human breast cancer cell lines inhibits proliferation, in vitro migration and in vivo tumor growth. Breast Cancer Res. 2011;13:1-14.
- [Google Scholar]
- MicroRNA-214 Suppresses The Growth and Invasiveness of Cervical Cancer Cells by Targeting UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 7. J Biol Chem. 2012;287:14301-9.
- [Google Scholar]
- MicroRNA-21 promotes cell transformation by targeting the programmed cell death 4 gene. Oncogene. 2008;27:4373-9.
- [Google Scholar]
- MicroRNA-21 (miR-21) post-transcriptionally downregulates tumor suppressor Pdcd4 and stimulates invasion, intravasation and metastasis in colorectal cancer. Oncogene. 2008;27:2128-36.
- [Google Scholar]
- BIC and miR-155 are highly expressed in Hodgkin, primary mediastinal and diffuse large B cell lymphomas. J Pathol. 2005;207:243-9.
- [Google Scholar]
- MicroRNA-based Cancer Therapeutics: Big Hope from Small RNAs. Mol Cell Pharmacol. 2010;2:213-9.
- [Google Scholar]
- Base pairing small RNAs and their roles in global regulatory networks. FEMS Microbiol Rev. 2010;34:866-82.
- [Google Scholar]
- Regulation by small RNAs in bacteria: expanding frontiers. Mol Cell. 2011;43:880-91.
- [Google Scholar]
- RNA polymerase III transcribes human microRNAs. Nat StructMolBiol. 2006;13:1097-101.
- [Google Scholar]
- Trans-splicing and polyadenylation of let-7 microRNA primary transcripts. RNA. 2004;10:1586-94.
- [Google Scholar]
- Processing of primary microRNAs by the Microprocessor complex. Nature. 2004;432:231-5.
- [Google Scholar]
- The Drosha-DGCR8 complex in primary microRNA processing. Genes Dev. 2004;18:3016-27.
- [Google Scholar]
- Exportin-5 mediates the nuclear export of premicroRNAs and short hairpin RNAs. Genes Dev. 2003;17:3011-6.
- [Google Scholar]
- Exportin 5 is a RanGTP-dependent dsRNAbinding protein that mediates nuclear export of pre-miRNAs. RNA. 2004;10:185-91.
- [Google Scholar]
- TRBP recruits the Dicer complex to Ago2 for microRNA processing and gene silencing. Nature. 2005;436:740-4.
- [Google Scholar]
- Human RISC couples microRNA biogenesis and posttranscriptional gene silencing. Cell. 2005;123:631-40.
- [Google Scholar]
- A human, ATP-independent, RISC assembly machine fueled by pre-miRNA. Genes Dev. 2005;19:2979-90.
- [Google Scholar]
- Large-scale sequencing reveals 21URNAsand additional microRNAs and endogenous siRNAs in C. elegans. Cell. 2006;127:1193-207.
- [Google Scholar]
- RISC is a 5’ phosphomonoester-producing RNA endonuclease. Genes Dev. 2004;18:975-80.
- [Google Scholar]
- mRNA degradation by miRNAs andGW182requires bothCCR4:NOTdeadenylase and DCP1:DCP2 decapping complexes. Genes Dev. 2006;20:1885-98.
- [Google Scholar]
- MicroRNAs control translation initiation by inhibiting eukaryotic initiation factor 4E/cap and poly(A) tail function. Proc Natl Acad Sci USA. 2005;102:16961-6.
- [Google Scholar]
- Short RNAs repress translation after initiation in mammalian cells. Mol Cell. 2006;21:533-42.
- [Google Scholar]
- MicroRNA-dependent localization of targeted mRNAs to mammalian P-bodies. Nat. Cell Biol. 2005;7:719-23.
- [Google Scholar]
- A crucial role for GW182 and the DCP1:DCP2 decapping complex in miRNA-mediated gene silencing. RNA. 2005;11:1640-47.
- [Google Scholar]
- Translation repression in human cells by microRNA-induced gene silencing requires RCK/p54. PLoS Biol. 2006;4:e210.
- [Google Scholar]
- A hexanucleotide element directs microRNA nuclear import. Science. 2007;315:97-100.
- [Google Scholar]
- Specificity of microRNA target selection in translational repression. Genes Dev. 2004;18:504-11.
- [Google Scholar]
- Natural selection on human microRNA binding sites inferred from SNP data. Nat Genet. 2006;38:1452-6.
- [Google Scholar]
- A pattern-based method for the identification of MicroRNA binding sites and their corresponding heteroduplexes. Cell. 2006;126:1203-17.
- [Google Scholar]
- Mechanisms of microRNA-mediated gene regulation in animal cells. Trends Genets. 2007;23:243-9.
- [Google Scholar]
- MicroRNA-10a binds the 5’UTR of ribosomal protein mRNAs and enhances their translation. Mol Cell. 2008;30:460-71.
- [Google Scholar]
- Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature. 2010;466:835-40.
- [Google Scholar]
- Cell-type-specific signatures of microRNAs on target mRNA expression. Proc Natl Acad Sci USA. 2006;103:2746-51.
- [Google Scholar]
- The let-7 MicroRNA family members mir-48, mir-84, and mir-241 function together to regulate developmental timing in Caenorhabditiselegans. Dev Cell. 2005;9:403-14.
- [Google Scholar]
- Drosophila lacking microRNA miR-278 are defective in energy homeostasis. Genes Dev. 2006;20:417-22.
- [Google Scholar]
- The Drosophila microRNA iab-4 causes a dominant homeotic transformation of halteres to wings. Genes Dev. 2005;19:2947-52.
- [Google Scholar]
- A pancreatic islet-specific microRNA regulates insulin secretion. Nature. 2004;432:226-30.
- [Google Scholar]
- A brain-specific microRNA regulates dendritic spine development. Nature. 2006;439:283-8.
- [Google Scholar]
- MicroRNAs regulate the expression of the alternative splicing factor nPTB during muscle development. Genes Dev. 2007;21:71-84.
- [Google Scholar]
- Serum response factor regulates a muscle-specific microRNA that targets Hand2 during cardiogenesis. Nature. 2005;436:214-20.
- [Google Scholar]
- Dysregulation of cardiogenesis, cardiac conduction, and cell cycle in mice lacking miRNA-1-2. Cell. 2007;129:303-17.
- [Google Scholar]
- Functional integration of microRNAs into oncogenic and tumor suppressor pathways. Cell Cycle. 2008;7:2493-9.
- [Google Scholar]
- Conservation of the sequence and temporal expression of let-7 heterochronic regulatory RNA. Nature. 2000;408:86-9.
- [Google Scholar]
- Unique microRNA molecular profiles in lung cancer diagnosis and prognosis. Cancer Cell. 2006;9:189-98.
- [Google Scholar]
- let-7 microRNA functions as a potential growth suppressor in human colon cancer cells. Biol Pharm Bull. 2006;29:903-6.
- [Google Scholar]
- The tumor suppressor microRNA let-7 represses the HMGA2 oncogene. Genes Dev. 2007;21:1025-30.
- [Google Scholar]
- MicroRNA let-7a down-regulates MYC and revertsMYCinduced growth in Burkitt lymphoma cells. Cancer Res. 2007;67:9762-70.
- [Google Scholar]
- Frequent deletions and down-regulation of micro-RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc Natl Acad Sci USA. 2002;99:15524-9.
- [Google Scholar]
- miR-15 and miR-16 induce apoptosis by targeting BCL2. Proc Natl Acad Sci USA. 2005;102:13944-9.
- [Google Scholar]
- Mir-29 regulates Mcl-1 protein expression and apoptosis. Oncogene. 2007;26:6133-40.
- [Google Scholar]
- MicroRNA signatures associated with cytogenetics and prognosis in acute myeloid leukemia. Blood. 2008;111:3183-9.
- [Google Scholar]
- microRNA-34a is tumor suppressive in brain tumors and glioma stem cells. Cell Cycle. 2010;9:1031-6.
- [Google Scholar]
- High expression of precursor microRNA-155/BIC RNA in children with Burkitt lymphoma. Genes Chrom Cancer. 2004;39:167-9.
- [Google Scholar]
- BIC and miR-155 are highly expressed in Hodgkin, primary mediastinal and diffuse large B cell lymphomas. J Pathol. 2005;207:243-9.
- [Google Scholar]
- A microRNA signature associated with prognosis and progression in chronic lymphocytic leukemia. N Engl J Med. 2005;353:1793-801.
- [Google Scholar]
- Pre-B cell proliferation and lymphoblastic leukemia/high-grade lymphoma in Eμ-miR155 transgenic mice. Proc Natl Acad Sci USA. 2006;103:7024-9.
- [Google Scholar]
- Requirement of BIC/microRNA-155 for normal immune function. Science. 2007;316:608-11.
- [Google Scholar]
- Extensive modulation of a set of microRNAs in primary glioblastoma. BiochemBiophys Res Commun. 2005;334:1351-8.
- [Google Scholar]
- MicroRNA-21 regulates expression of the PTEN tumor suppressor gene in human hepatocellular cancer. Gastroenterology. 2007;133:647-58.
- [Google Scholar]
- Programmed cell death 4 (PDCD4) is an important functional target of the microRNA miR-21 in breast cancer cells. J Biol Chem. 2008;283:1026-33.
- [Google Scholar]
- MicroRNA-21 targets the tumor suppressor gene tropomyosin 1 (TPM1) J Biol Chem. 2007;282:14328-36.
- [Google Scholar]
- Identification and characterization of a novel gene,C13orf25, as a target for 13q31-q32 amplification in malignant lymphoma. Cancer Res. 2004;64:3087-95.
- [Google Scholar]
- miRiad roles for the miR-17-92 cluster in development and disease. Cell. 2008;133:217-22.
- [Google Scholar]
- Transgenic overexpression of the microRNA miR-17-92 cluster promotes proliferation and inhibits differentiation of lung epithelial progenitor cells. DevBiol. 2007;310:442-53.
- [Google Scholar]
- Lymphoproliferative disease and autoimmunity in mice with elevated miR-17-92 expression in lymphocytes. Nat Immunol. 2008;9:405-14.
- [Google Scholar]
- Myc and Ras collaborate in inducing accumulation of activecyclin E/Cdk2 and E2F. Nature. 1997;387:422-6.
- [Google Scholar]
- E2F1-regulated microRNAs impair TGFβ-dependent cell cycle arrest and apoptosis in gastric cancer. Cancer Cell. 2008;13:272-86.
- [Google Scholar]
- miR-17-5p regulates breast cancer cell proliferation by inhibiting translation of AIB1 mRNA. Mol Cell Biol. 2006;26:8191-201.
- [Google Scholar]
- Differentiation-associated miR-22 represses Max expression and inhibits cell cycle progression. Biochem Biophys Res Commun. 2010;394:606-11.
- [Google Scholar]
- A Genetic Screen Implicates miRNA-372 and miRNA-373 As Oncogenes in Testicular Germ Cell Tumors. Cell. 2006;124:1169-81.
- [Google Scholar]
- MicroRNA gene expression deregulation in human breast cancer. Cancer Res. 2005;65:7065-70.
- [Google Scholar]
- Antisense inhibition of human miRNAs and indications for an involvement of miRNA in cell growth and apoptosis. Nucleic Acids Res. 2005;33:1290-7.
- [Google Scholar]
- Altered miRNA expression in sputum for diagnosis of non-small cell lung cancer. Lung cancer. 2010;67:170-6.
- [Google Scholar]
- Reduced accumulation of specific microRNAs in colorectal neoplasia. Mol Cancer Res. 2003;1:882-91.
- [Google Scholar]
- Early detection of lung adenocarcinoma in sputum by a panel of microRNA markers. Int J Cancer. 2010;127:2870-8.
- [Google Scholar]
- MicroRNAs and cancer therapy: The next wave or here to stay? Cancer Biol Ther. 2010;9:479-82.
- [Google Scholar]
- Molecular profiling uncovers a p53-associated role for microRNA-31 in inhibiting the proliferation of serous ovarian carcinomas and other cancers. Cancer Res. 2010;70:1906-15.
- [Google Scholar]
- miR-17-92 expression in differentiated T cells - implications for cancer immunotherapy. J Transl Med. 2010;8:17. doi:10.1186/1479-5876-8-17
- [Google Scholar]
- MiR-15a and miR-16-1 cluster functions in human leukemia. Proc Natl Acad Sci USA. 2008;105:5166-71.
- [Google Scholar]
- Tumor-suppressive miR-34a induces senescence-like growth arrest through modulation of the E2F pathway in human colon cancer cells. ProcNatlAcadSci USA. 2007;104:15472-7.
- [Google Scholar]
- The let-7 microRNA reduces tumor growth in mouse models of lung cancer. Cell Cycle. 2008;7:759-64.
- [Google Scholar]
- Widespread microRNA repression by Myc contributes to tumorigenesis. Nat Genet. 2008;40:43-50.
- [Google Scholar]
- Therapeutic silencing of miR-10b inhibits metastasis in a mousemammary tumor model. Nat Biotechnol. 2010;28:341-7.
- [Google Scholar]
- Antagonism of microRNA-122 in mice by systemically administered LNA-antimiR leads to up-regulation of a large set of predicted target mRNAs in the liver. Nucleic Acids Res. 2008b;36:1153-62.
- [Google Scholar]
- The promises and pitfalls of RNA-interference based therapeutics. Nature. 2009;457:426-33.
- [Google Scholar]
- Delivery of MicroR A-10b with olylysineanoparticles for Inhibition of Breast cancer ell Wound Healing. Breast Cancer (Auckl). 2012;6:9-19.
- [Google Scholar]
- Nanoparticles Modified With Tumor-targeting scFv Deliver siRNA and miRNA for Cancer Therapy. Mol Ther. 2005;189:1650-6.
- [Google Scholar]
- PLGA-based gene delivering nanoparticle enhance suppression effect of miRNA in HePG2 cells. Nanoscale Research Letters. 2011;6:447-55.
- [Google Scholar]
- Molecular imaging of a cancer-targeting theragnostics probe using a nucleolinaptamer- and microRNA-221 molecular beacon-conjugated nanoparticle. Biomaterials. 2012;33:207-17.
- [Google Scholar]
- Effective delivery of anti-miRNA DNA oligonucleotides by functionalized gold nanoparticles. Journal of Biotechnology. 2011;155:287-92.
- [Google Scholar]
- CdTe nanoparticles display tropism to core histones and histone-rich cell organelles. Small. 2008;4:2006-15.
- [Google Scholar]
- Quantum dot-induced epigenetic and genotoxic changes in human breast cancer cells. J Mol Med. 2008;86:291-302.
- [Google Scholar]
- Unmodified cadmium telluride quantum dots induce reactive oxygen species formation leading to multiple organelle damage and cell death. Chem Biol. 2005;12:227-34.
- [Google Scholar]
- MicroRNAs as participants in cytotoxicity of CdTe quantum dots in NIH/3T3 cells. Biomaterials. 2011;32:3807-14.
- [Google Scholar]
- Targeted delivery of antisense inhibitor of miRNA for antiangiogenesis therapy using cRGD-functionalized nanoparticles. Mol Pharm. 2011;8:250-9.
- [Google Scholar]
- Potent and specific genetic interference by double-stranded RNA in Caenorhabditiselegans. Nature. 1998;391:806-11.
- [Google Scholar]






