Translate this page into:
Streptococcus pseudopneumoniae: an emerging respiratory tract pathogen
Reprint requests: Dr B. Dhanashree, Associate Professor, Department of Microbiology, Kasturba Medical College, Manipal University, Mangalore 575 001, India e-mail: dbiranthabail@yahoo.co.in
-
Received: ,
This is an open-access article distributed under the terms of the Creative Commons Attribution-Noncommercial-Share Alike 3.0 Unported, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
This article was originally published by Medknow Publications & Media Pvt Ltd and was migrated to Scientific Scholar after the change of Publisher.
Abstract
Background & objectives:
Streptococcus pseudopneumoniae a member of the Viridans Streptococci, is known to be associated with chronic obstructive pulmonary disease and respiratory tract infections (RTI). Very scanty information is available on the isolation of S. pseudopneumoniae from India. Hence, the present study was an attempt to isolate S. pseudopneumoniae from clinical samples and to study their drug resistance pattern.
Methods:
Sputum samples (n=150) submitted to the microbiology laboratory for routine culture from patients clinically suspected to have lower respiratory tract infection were inoculated onto sheep blood agar and chocolate agar plates. Alpha haemolytic colonies were identified as S. pseudopneumoniae based on absence of capsule, bile solubility and optochin susceptibility in 5 per cent CO2 and ambient air. Disk diffusion method was used for antibiotic susceptibilily testing.
Results:
Among the samples screened, 4 per cent showed the growth of only S. pseudopneumoniae. Other pathogens isolated were Streptococcus pneumoniae, Moraxella catarrhalis, Klebsiella spp., Enterococcus spp., Pseudomonas spp., Haemophilus influenzae, Staphylococcus aureus, Candida albicans. All the S. pseudopneumoniae isolates were resistant to erythromycin.
Interpretation & conclusions:
Our preliminary results showed presence of S. pseudopneumoniae in this part of the country and these were associated with RTI. Currently, most clinical laboratories report optochin susceptible isolates in 5 per cent CO2 as S. pneumoniae and the resistant ones are not further tested for susceptibility in ambient air. As a result, S. pseudopneumoniae may be missed out. Hence, performance of at least two tests, viz. optochin susceptibility with incubation in 5 per cent CO2 and ambient air along with bile solubility is necessary to differentiate S. pneumoniae from S. pseudopneumoniae
Keywords
Antibiotic susceptibility
purulent sputum
respiratory tract pathogen
Streptococcus pseudopneumoniae
Streptococcus pseudopneumoniae is a recently described member of the Streptococcus mitis/oralis group of viridans Streptococci that shares some characteristics with S. pneumoniae1. Key features of S. pseudopneumoniae are the absence of pneumococcal capsule, insolubility in bile, resistance or indeterminate susceptibility to optochin when incubated in 5 per cent CO2 but susceptibility to optochin when incubated in ambient air, positive reactions in DNA probe hybridization and antigen detection tests12. Currently, most clinical laboratories in India depend only on the optochin susceptibility test done in 5 per cent CO2 but not in ambient air for S. pneumoniae identification. As a result, S. pseudopneumoniae may be missed out or wrongly identified.
The clinical relevance of S. pseudopneumoniae has not yet been fully established, although it is found to be associated with chronic obstructive pulmonary disease2. Very few reports are available on potential pathogenic effect and drug resistance of S. pseudopneumoniae3–5. Until now, perhaps there are no published reports on the isolation of S. pseudopneumoniae from clinical samples in India. Therefore, an attempt was made to isolate S. pseudopneumoniae from sputum samples collected from patients suspected to have lower respiratory tract infection (LRTI) and to study their antibiogram.
Material & Methods
Collection of samples: Sputum samples (n=200) of patients, clinically suspected of having LRTI, submitted to the microbiology department of Kasturba Medical College, Mangalore, Karnataka, India, during March 2010 to August 2010 for routine culture and sensitivity testing were included in the study by following convenient sampling method.
Microscopic examination: Gram stain of sputum samples (n=150) showing >25 leukocytes and <10 squamous epithelial cells/×100 microscopic field along with large number of Gram positive cocci in pairs were included in the study and processed further for culture. Remaining sputum samples (n=50) showing >10 epithelial cells/x100 microscopic field were excluded from the study.
Culture and identification of the isolates: Samples were inoculated onto 5 per cent sheep blood agar and chocolate agar plates and incubated at 37°C for 24 h. Alpha haemolytic colonies on the culture plates, that were Gram positive cocci in pairs by staining, were identified as S. pseudopneumoniae and differentiated from S. pneumoniae on the basis of the tests for pneumococcal capsule, bile solubility and optochin susceptibility2.
Bile solubility test was performed according to standard procedures5. Capsules were detected by observing a halo around Pneumococci with India ink at × 400 magnification3.
Optochin susceptibility test: Colonies from overnight cultures were inoculated onto 5 per cent sheep blood agar plates in duplicate. Optochin disks of 6 mm diameter containing 5 μg antibiotic [Hi-media Laboratories (P) Ltd., Mumbai] were placed on both the inoculated blood agar plates. One plate was incubated in 5 per cent CO 2and another in ambient air (O2 atmosphere) at 37°C for 24 h. Zones of inhibition around the disk were measured and interpreted as per Clinical and Laboratory Standards Institute (CLSI) guidelines36. S. pneumoniae strain ATCC 49619 procured from Himedia Laboratories (P) Ltd., Mumbai, was used as control. Isolates were considered to be resistant to optochin when the inhibition zones were smaller than 14 mm.
Antibiotic susceptibility test: Biochemically confirmed S. pseudopneumoniae were subjected to antibiotic susceptibility testing on Mueller Hinton blood agar plates by Kirby Bauer's disk diffusion method7 using antibiotics like cefepime, oxacillin, cefotaxime, ciprofloxacin, chloramphenicol, erythromycin, meropenem, tetracycline, teicoplanin, trimethoprim-sulphamethoxazole and vancomycin. Plates were incubated at 37°C for 24 h. The result was interpreted as either sensitive, intermediate or resistant as per CLSI guidelines6. S.pneumoniae strain ATCC 49619 was used as control.
Results & Discussion
Of the 150 sputum samples cultured, 25 showed the growth of α haemolytic streptococci. Among these, six isolates lacked capsules, were insoluble in 10 per cent bile, showed resistance to optochin (zone size <14 mm diameter when incubated in 5% CO2) but were susceptible to optochin (zone sizes between 18 to 25 mm diameter) when incubated in ambient air. Hence, these six isolates were presumptively identified as S. pseudopneumoniae biochemically. S. pseudopneumoniae colonies grown on 5 per cent sheep blood agar were typically small (up to 1 mm in diameter after 24 h of incubation), smooth, shiny, and domed, with entire edges. Occasional colonies had depressed centers, appearing as a smaller version of the draftsmen colonies of S. pneumoniae.
The remaining 19 α-haemolytic Streptococci were identified as S. pneumoniae due to the presence of capsule, bile soluble and sensitive to optochin at 5 per cent CO2 and generally poor growth or no growth in ambient air. The colonies had depressed centers typical of S. pneumoniae. Remaining 125 samples, showed heavy growth of various pathogens like Moraxella catarrhalis (n=21), Klebsiella spp. (n=8), Enterococcus spp. (n=6), Pseudomonas spp. (n=5), Haemophilus influenzae (n=6), Staphylococcus aureus (n=4) and Candida albicans (n=10). Some of the samples (n=35) also showed scanty growth of other Streptococcus spp. (Streptococci which were biochemically neither S. pneumoniae nor S. pseudopneumoniae) and mixed scanty growth of oropharyngeal flora (n=30).
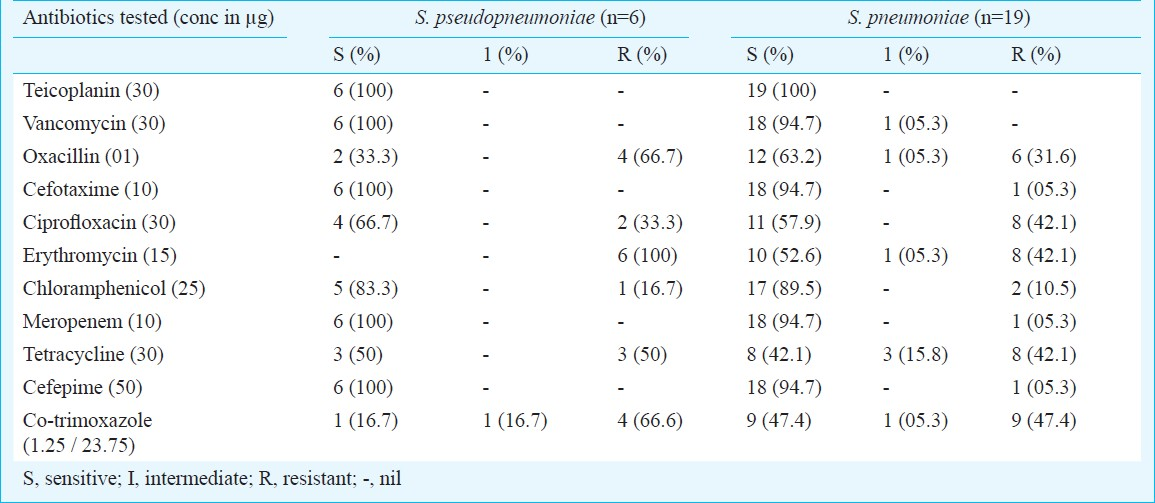
All six S. pseudopneumoniae isolates, were sensitive to teicoplanin, vancomycin, cefotaxime, meropenem, cefepime and showed resistance to erythromycin. S. pseudopneumoniae isolates sensitive to ciprofloxacin and tetracycline were found to be four and three, respectively. Resistance to penicillin (as determined by oxacillin screening test) and co-trimoxazole was 66 per cent. All S. pneumoniae isolates were sensitive to teicoplanin (Table).
Our findings indicated the presence of S. pseudopneumoniae in this region which could be isolated from a small proportion of sputum samples. All six isolates of S. pseudopneumoniae were the predominant or only microorganism from good-quality purulent sputum samples obtained from patients with symptomatic LRTI and not from URTI. Further, in all six cases, the sputum Gram stain results indicated the presence of Gram positive cocci in pairs as the predominant bacteria along with pus cells. These findings provide supporting evidence of a potential pathogenic role of S. pseudopneumoniae.
All six S. pseudopneumoniae isolates were found to be resistant to erythromycin, four were resistant to penicillin (by oxacillin screening test) and co-trimoxazole, three were resistant to tetracycline and two to ciprofloxacin. Our findings corroborated with those reported from New Zealand and North America24. We could not find any reported cases of isolation of S. pseudopneumoniae from India to compare our results. However, antibiotic resistance in these isolates needs to be confirmed by detecting the minimum inhibitory concentration (MIC) for the resistant isolates.
High prevalence of antimicrobial resistance in major respiratory pathogens has led to serious concern in the selection of an appropriate antimicrobial agent for the empirical treatment of respiratory tract infections worldwide. Increasing penicillin and multidrug resistance in S. pneumoniae has important clinical implications. In the present study, resistance to penicillin as determined by oxacillin screening test was found to be 31 per cent and that of tetracycline and co-trimoxazole were found to range from 42 to 47 per cent for S. pneumoniae. These results are in agreement with the previous findings8. However, resistance to erythromycin and ciprofloxacin was also found.
Currently, most clinical laboratories depend only on the optochin susceptibility test by incubation in 5 per cent CO2 for identification of S. pneumoniae. If the clinical sample has S. pseudopneumoniae it will show resistance to optochin when incubated only in 5 per cent CO2 and such isolates are likely to be reported as oropharyngeal flora. Though S. pseudopneumoniae have been isolated in this region from sputum samples of patients with LRTI, further studies are necessary to prove the potential pathogenic role of these bacteria in lower respiratory tract infection or in chronic obstructive pulmonary disease (COPD). Since the presumptive identification of S. pseudopneumoniae was done by biochemical reaction, further characterization of these isolates by molecular methods is necessary. Testing of large number of samples, increased awareness to look for the presence of S. pseudopneumoniae and characterization to the genetic level will help to better determine its prevalence and clinical importance. Preliminary data from this study should prompt further research to characterize the role of S. pseudopneumoniae in COPD and LRTI.
Acknowledgment
Sariya thanks Indian Council of Medical Research (ICMR), New Delhi, for the award of Short-Term Research Studentship (STS) for this study.
References
- Accuracy of phenotypic and genotypic testing for identification of Streptococcus pneumoniae and description of Streptococcus pseudopneumoniae sp.nov. J Clin Microbiol. 2004;42:4686-96.
- [Google Scholar]
- Characteristics of Streptococcus pseudopneumoniae isolated from purulent sputum samples. J Clin Microbiol. 2006;44:923-7.
- [Google Scholar]
- Incidence and pathogenic effect of Streptococcus pseudopneumoniae. J Clin Microbiol. 2006;44:2240-1.
- [Google Scholar]
- Antimicrobial susceptibility profile of Streptococcus pseudopneumoniae isolated from sputum. Antimicrob Agents Chemother. 2008;52:2998.
- [Google Scholar]
- Streptococcus. In: Murray PR, Baron EJ, Jorgensen JH, Pfaller MA, Yolken RH, eds. Manual of clinical microbiology (8th ed). Washington DC: American Society for Microbiology; 2003. p. :405-21.
- [Google Scholar]
- Clinical and Laboratory Standards Institute (CLSI) In: Performance standards for antimicrobial susceptibility testing. Wayne, Pa: CLSI; 2007. M100-S17
- [Google Scholar]
- Antibiotic susceptibility testing by a standaridized disc method. Am J Clin Pathol. 1996;45:493-6.
- [Google Scholar]
- Antimicrobial resistance in invasive and colonising Streptococcus pneumoniae in North India. Indian J Med Microbiol. 2007;25:256-9.
- [Google Scholar]






