Translate this page into:
Molecular strain typing of Trichophyton mentagrophytes (T. mentagrophytes var. interdigitale) using non-transcribed spacer region as a molecular marker
Reprint requests: Dr Anupma Jyoti Kindo, Department of Microbiology, Sri Ramachandra University, Porur, Chennai 600 116, Tamil Nadu, India e-mail: anupmalakra@gmail.com
-
Received: ,
This is an open access article distributed under the terms of the Creative Commons Attribution-NonCommercial-ShareAlike 3.0 License, which allows others to remix, tweak, and build upon the work non-commercially, as long as the author is credited and the new creations are licensed under the identical terms.
This article was originally published by Medknow Publications & Media Pvt Ltd and was migrated to Scientific Scholar after the change of Publisher.
Abstract
Background & objectives:
Dermatophytes are keratinophilic fungi that infect keratinized tissues of human and animal origin. Trichophyton mentagrophytes is considered to be a species complex composed of several strains, which include both anthropophiles and zoophiles. Accurate discrimination is critical for comprehensive understanding of the clinical and epidemiological implications of the genetic heterogeneity of this complex. Molecular strain typing renders an effective way to discriminate each strain. The objective of the study was to characterize T. mentagrophytes clinical isolates to sub-species level using molecular techniques and non-transcribed spacer (NTS) region as marker.
Methods:
Sixty four T. mentagrophytes clinical isolates were identified by phenotypic methods. These were subjected to polymerase chain reaction targeting three sub-repeat elements (SREs), TmiS0, TmiS1 and TmiS2 of the NTS region. Sequence analysis of internal transcribed spacer (ITS) region of different types was also done.
Results:
Strain-specific polymorphism was observed in all three loci. Totally, 13 different PCR types were obtained on combining all the three SREs loci. No variation was observed in the ITS region.
Interpretation & conclusions:
The study described the usefulness of molecular strain typing technique for the discrimination of the T. mentagrophytes isolates. This will help for the future explorations into the epidemiology of T. mentagrophytes and its complex.
Keywords
Dermatophytes
molecular strain typing
non-transcribed spacer region
polymerase chain reaction
sub-repeat elements
Trichophyton mentagrophytes var. interdigitale
Dermatophytes are the most common agents of superficial mycoses. Dermatophytes are adapted to utilize keratin as a major nutritional source; hence, infection is generally cutaneous and restricted to the non-living cornified layers such as skin, stratum corneum, hair and nails of humans and animals1. In India, Trichophyton mentagrophytes is next to Trichophyton rubrum in causing dermatophytosis23. T. mentagrophytes is both anthropophilic and zoophilic. The downy form is associated with chronic human infections, whereas the granular form causes animal and acute human infections4.
Although T. rubrum and T. mentagrophytes are well separated in phylogeny, the clinical conditions manifested are similar. These show considerable variation in microscopic and cultural characteristics, but these cannot be taken as strain markers as the phenotypic characters of T. mentagrophytes can change notably on routine sub-culture.
Genotypic approaches have been proven to be useful for solving problems in dermatophyte taxonomy, as well as enhancing the reliability and speed of dermatophytosis diagnosis and strain differentiation5. Genotyping using variable tandem sub-repeat elements (SREs) of non-transcribed spacer (NTS) region in the ribosomal DNA (rDNA) gene cluster has been demonstrated in T. rubrum6. T. mentagrophytes var. interdigitale has also been shown to possess genetic polymorphisms that map to the rDNA789. The NTS region in T. mentagrophytes var. interdigitale consists of three highly repetitive SREs, namely TmiS0, TmiS1 and TmiS2. These regions are more prone to mutations due to unequal crossing over between strains and have been used to determine the intra-species variations89. Identifying the isolates to the strain level along with clinical details will aid in tracking the source of infection. Only a few studies have been conducted on strain typing of T. mentagrophytes89 in other countries, which includes representative Indian clinical isolates. The present study was therefore, conducted to characterize T. mentagrophytes isolates to sub-species level using molecular strain typing techniques and using NTS region as a molecular marker.
Material & Methods
Sixty four T. mentagrophytes isolates received from April 2012 to March 2014 in the Mycology division, department of Microbiology, Sri Ramachandra Medical College & Research Institute, Chennai, India, were used in this study. All these isolates were characterized based on macroscopic and microscopic appearance from the primary culture.
Phenotypic characterization of T. mentagrophytes: All isolates were grown in Sabouraud dextrose agar (SDA) (Hi-Media, Mumbai) with actidione. The culture characteristics were observed on SDA and dermatophyte test medium (Hi-Media). Microscopic characteristics were observed in lactophenol cotton blue (LPCB) (Hi-Media) mount. Those isolates which did not demonstrate proper microscopic characters in LPCB mount were subjected to slide culture technique with oatmeal agar (Hi-Media) and observed. The isolates which were confirmed as T. mentagrophytes were taken for molecular characterization.
Molecular characterization of T. mentagrophytes
DNA isolation: DNA was extracted from all the 64 T. mentagrophytes isolates by boiling method10, with certain modifications. Briefly, the culture was suspended in 400 μl lysis buffer [10 mM TRIS, (pH 8), 1 mM EDTA (pH 8), 3% SDS and 100 mM NaCl] in a 1.5 ml microfuge tube; 20 μl of proteinase K (1 mg/ml) (Merck GeNei, India) was added and incubated at 56°C for 30 min. It was boiled for one minute and 400 μl of phenol:chloroform (Sigma, USA) (1:1) mixture was added, mixed well and centrifuged at 8600 g for 10 min. The aqueous layer was transferred to a new microfuge tube; equal volume of chloroform was added, mixed well and centrifuged at 8600 g for 10 min. The aqueous layer was transferred to a new microfuge tube. DNA was precipitated using equal volume of ice cold isopropyl alcohol and washed twice with 70 per cent ethanol. The pellet was suspended in 40 μl sterile nuclease-free water and stored at −20°C until use.
Amplification of non-transcribed spacer (NTS) region: PCR amplification of all the three SREs was performed individually in an Eppendorf Gradient Mastercycler (Eppendorf, Germany). Primer sequences used for all three regions (sub-repeat regions, S0, S1 and S2) are listed in (Table I). Each reaction mix contained 25 μl PCR master mix (Merck GeNei), 50 pmol forward (Sigma) and reverse primers (Sigma) each, and 1 μl DNA template. The volume was made up to 50 μl with nuclease-free water (Merck GeNei).

PCR was carried out for all three sub-repeat regions under the same reaction conditions with denaturation at 95°C for 30 sec, annealing at 55°C for 30 sec and extension at 72°C for 90 sec. The cycle was repeated 35 times. Further, an initial denaturation at 95°C for five minutes, and final extension at 72°C for 10 min were added.
Amplification of internal transcribed spacer (ITS) region: The reaction mix contained 25 μl PCR master mix (Merck GeNei), 50 pmol universal fungal primers, ITS-1 (Sigma) and ITS-4 (Sigma) each, 1 μl of template DNA and the volume made up to 50 μl with nuclease-free water. Amplification was carried out under the following conditions: initial denaturation at 95°C for five minutes, denaturation at 95°C for 30 sec, annealing at 56°C for 30 sec, extension at 72°C for 30 sec and final extension at 72°C for five minutes.
Agarose gel electrophoresis: All three SREs PCR products and ITS products were electrophoresed in 1.5 per cent agarose gel in tris-acetate-EDTA buffer, stained with ethidium bromide (0.5 μg/ml), visualized under ultraviolet light and photographed.
Gene sequencing: The ITS region was sequenced for representative isolates of each NTS type (SciGenom Labs, Cochin, Kerala, India). The sequences were then used for nucleotide-nucleotide search using the BLAST algorithm at the NCBI website (http://www.ncbi.nlm.nih.gov/BLAST/). BLAST hits more than 98 per cent were considered.
Results
Phenotypic characterization of T. mentagrophytes: The colonies were powdery (39/64) to floccose (25/64), cream to yellowish buff coloured. Microconidia were spherical, sessile, arranged in dense, grape-like clusters or along the hyphae. The spiral hyphae were present in 52 of 64 isolates (81%). Macroconidia were 3-8 celled, smooth and thin-walled, clavate to cigar-shaped.
Molecular characterization of T. mentagrophytes: All three SREs, TmiS0, TmiS1 and TmiS2 were amplified and each region produced different banding patterns. As banding patterns were not identical with the previous studies89, all three SREs types were designated numerically and the previous study's nomenclature was not followed.
The PCR which targeted TmiS0 loci produced six different banding patterns with varying product size of approximately 180-800 bp (Fig. A). Two to four bands were observed in each type. The isolates were sorted based on the banding patterns and tabulated (Table II). Among them, type 2 was predominant accounting for a total of 46 of 64 isolates (71.8%). PCR which amplified TmiS1 loci produced five patterns with two to four bands in each type. The product size ranged approximately 180-1100 bp (Fig. B). Type 2 dominated with a total of 50 of the 64 isolates (78.1%) and the types are described in Table II. PCR amplification of TmiS2 loci produced three banding patterns with maximum of two bands. The types are tabulated in Table II. The product size ranged from 280 to 680 bp (Fig. C). Among them, type 3 was predominant with a total of 57 of the 64 isolates (89%).
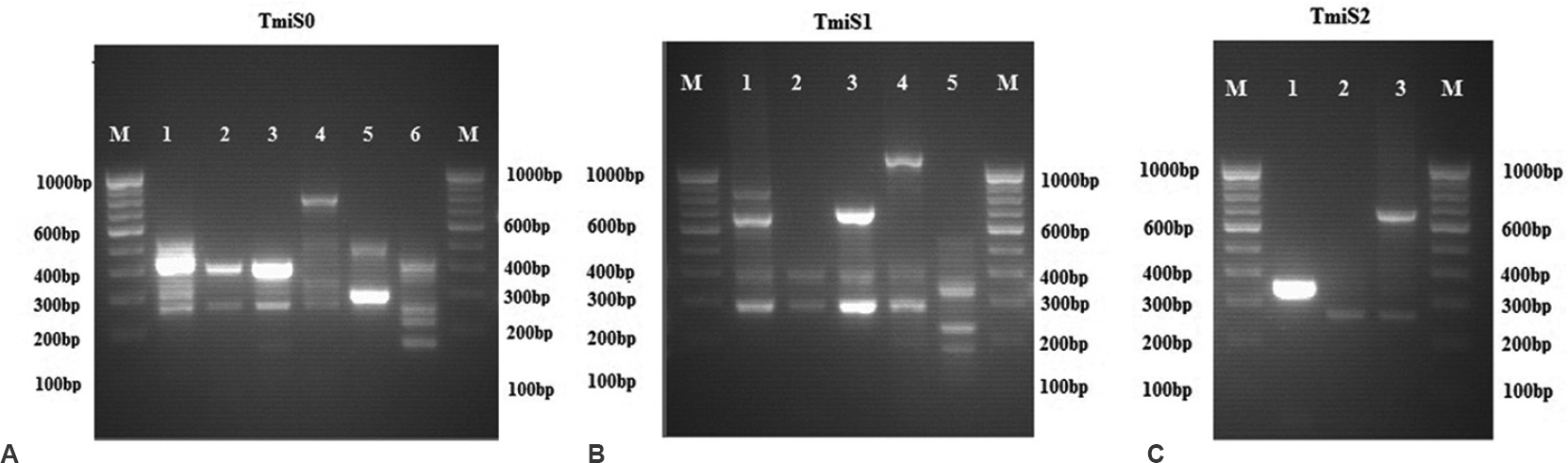
- Polymerase chain reaction fingerprint of representative types of Trichophyton mentagrophytes TmiS0 (A), TmiS1 (B) and TmiS2 (C) loci.

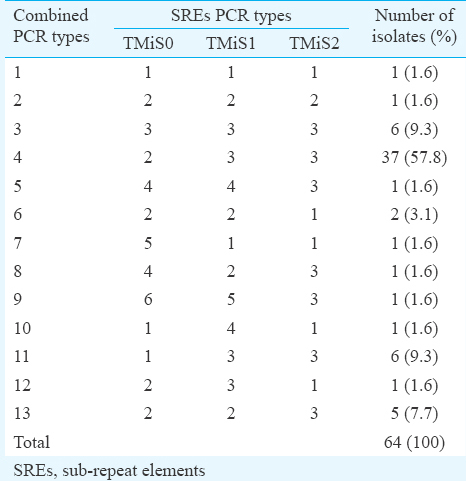
The variation between the three SREs (TmiS0, TmiS1 and TmiS2) was determined and grouped based on the differences in the banding pattern (Table II). The variation in the whole NTS region was studied by combining the banding patterns of TmiS0, TmiS1 and TmiS2 loci (Table III). The variability in banding pattern produced by individual locus appeared to occur independently. When all the three PCR patterns were combined, a total of 13 types were observed from the 64 isolates.
Internal transcribed spacer (ITS) sequence: All the 64 isolates were phenotypically confirmed as T. mentagrophytes. Of the 13 different NTS types observed in this study, ITS region of one representative isolate from each type was amplified and sequenced to confirm the isolates genotypically. BLAST hits showed more than 99 per cent identity to T. mentagrophytes var. interdigitale. The sequences were deposited in Genbank database (Accession numbers KP099590-KP099602).
Discussion
Large and small unit ribosomal RNA genes are arranged in clusters as tandem repeats in multiple copies. The tandem repeats are flanked both sides by NTS region which is more prone to mutation than the transcribed regions. Hence, the NTS region can be utilized for typing of dermatophytes to sub-species level. Earlier studies on T. rubrum6111213, T. tonsurans1415161718 and T. mentagrophytes var. interdigitale89 have used NTS region for investigating polymorphism and sub-species level discernments. In the current study, the primers for NTS used to type T. mentagrophytes var. interdigitale in the UK and Japan89, were utilized. As there was no similarity in banding patterns with previous studies, all three PCR types were named numerically to avoid overlap in pattern names. The TmiS0 locus had the greatest polymorphism by producing most number of PCR types among all three loci. Of the six types observed, band size 400 bp was shared by four types which accounted for almost 61 of 64 isolates (94%) but none of the isolate produced single band.
The TmiS1 locus also had five PCR types with multiple bands. Bands sized 300 and 400 bp were shared by type 1-4 which accounted for 63 of 64 isolates (98%). The TmiS1 locus was the longest and produced band of size 1100 bp. As demonstrated in earlier studies89, TmiS2 locus was the least polymorphic region which produced only three types in which 280 bp sized band was shared by two types accounting 58 of 64 isolates (91%).
Conjoining all three loci in NTS region produced a total of 13 types among 64 isolates, of which 37 (57.8%) were type 2-3-3, six (9.3%) of each type 3-3-3 and 1-3-3, five (7.7%) were 2-2-3, two (3.1%) were 2-2-1 and other eight types were produced by single isolates. The study performed in the UK failed to amplify TmiS0 and TmiS1 loci8, whereas in our study all three loci were amplified. None of the isolates produced patterns identical to those described in the UK or Japanese study89. The size of the PCR products from all three SREs was also very smaller than that observed in the earlier studies89. This could be due to degeneration in NTS region in Indian isolates compared to the UK and Japan strains89. Further investigations need to be done to examine the differences. The ITS region of one isolate from each of 13 NTS types was sequenced and analyzed for polymorphism. There was no difference observed in the ITS 1 or ITS 2 region. This showed that ITS region had less polymorphism compared to NTS region, making the latter one to be highly sensitive.
In conclusion, our study confirmed the molecular strain typing method as a reliable and suitable for sub-species level discernment of T. mentagrophytes var. interdigitale. This method can be adopted as a valuable tool for future explorations into the epidemiology of T. mentagrophytes complex and to track the source of infection.
Conflicts of Interest: None.
References
- Markers for host-induced gene expression in Trichophyton dermatophytosis. Infect Immun. 2005;73:6584-90.
- [Google Scholar]
- Trichophyton rubrum - The predominant etiological agent in human dermatophytosis in Chennai, India. Afr J Microbiol Res. 2007;1:9-12.
- [Google Scholar]
- Epidemiology of dermatophytosis in and around Tiruchirapalli, Tamilnadu, India. Asian Pac J Trop Dis. 2012;2:286-9.
- [Google Scholar]
- Natural infection in laboratory animals due to Trichophyton mentagrophytes in India. Mycopathol Mycol Appl. 1964;24:275-80.
- [Google Scholar]
- Strain identification of Trichophyton rubrum by specific amplification of subrepeat elements in the ribosomal DNA nontranscribed spacer. J Clin Microbiol. 2000;38:4527-34.
- [Google Scholar]
- Restriction fragment length polymorphism analysis of ribosomal DNA intergenic regions is useful for differentiating strains of Trichophyton mentagrophytes. J Clin Microbiol. 2003;41:4583-8.
- [Google Scholar]
- PCR fingerprinting of Trichophyton mentagrophytes var. interdigitale using polymorphic subrepeat loci in the rDNA nontranscribed spacer. J Clin Microbiol. 2006;55:1349-55.
- [Google Scholar]
- Molecular typing of Trichophyton mentagrophytes var. interdigitale isolated in a university hospital in Japan based on the non-transcribed spacer region of the ribosomal RNA gene. J Dermatol. 2010;37:431-40.
- [Google Scholar]
- Molecular species identification of Candida from blood samples of Intensive Care Unit patients by polymerase chain reaction - restricted fragment length polymorphism. J Lab Physicians. 2012;4:1-4.
- [Google Scholar]
- PCR typing of Trichophyton rubrum isolates by specific amplification of subrepeat elements in ribosomal DNA nontranscribed spacer. Iran J Dermatol. 2008;11:17-20.
- [Google Scholar]
- Single strains of Trichophyton rubrum in cases of tinea pedis. J Med Microbiol. 2005;54 (Pt 8):725-6.
- [Google Scholar]
- Molecular typing of Trichophyton rubrum clinical isolates from Poland. Mycoses. 2011;54:e726-36.
- [Google Scholar]
- Genetic heterogeneity in the rRNA gene locus of Trichophyton tonsurans. J Clin Microbiol. 2003;41:5478-87.
- [Google Scholar]
- Molecular typing of Trichophyton tonsurans by PCR-RFLP of the ribosomal DNA nontranscribed spacer region. J Dermatol Sci. 2004;36:125-7.
- [Google Scholar]
- Genotype analysis of the variable internal repeat region in the rRNA gene of Trichophyton tonsurans isolated from Japanese Judo practitioners. Microbiol Immunol. 2006;50:57-60.
- [Google Scholar]
- Molecular epidemiology of Trichophyton tonsurans isolated in Japan using RFLP analysis of non-transcribed spacer regions of ribosomal RNA genes. Jpn J Infect Dis. 2007;60:188-92.
- [Google Scholar]
- Epidemiology of sporadic (non-epidemic) cases of Trichophyton tonsurans infection in Japan based on PCR-RFLP analysis of non-transcribed spacer region of ribosomal RNA gene. Jpn J Infect Dis. 2008;61:219-22.
- [Google Scholar]






