Translate this page into:
Genome-wide scan for hypertension linkage to chromosome 12q23.1 - q23.3 in a Chinese family
Reprint requests: Dr Shijie Xu, Chinese National Human Genome Center at Shanghai, Shanghai 201203, PR China e-mail: xusj@chgc.sh.cn / Dr Gengru Jiang, Xinhua Hospital, Shanghai Jiaotong University School of Medicine, Shanghai 200092, PR China e-mail: jianggeng-ru@hotmail.com
-
Received: ,
This is an open-access article distributed under the terms of the Creative Commons Attribution-Noncommercial-Share Alike 3.0 Unported, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
This article was originally published by Medknow Publications & Media Pvt Ltd and was migrated to Scientific Scholar after the change of Publisher.
Abstract
Background & objectives:
Essential hypertension is a multifactorial disorder with a complex phenotype. Here we report a susceptibility locus for the hypertension mapped by a genome-wide microsatellite scanning in an affected Chinese family, in which 11 members had hypertension before the age of 40.
Methods:
A total of 22 individuals from a single family from Shanghai, PR China, were genotyped on more than 400 microsatellite markers with a spacing gap of less than 10 cm for nearly the entire scanned genome.
Results:
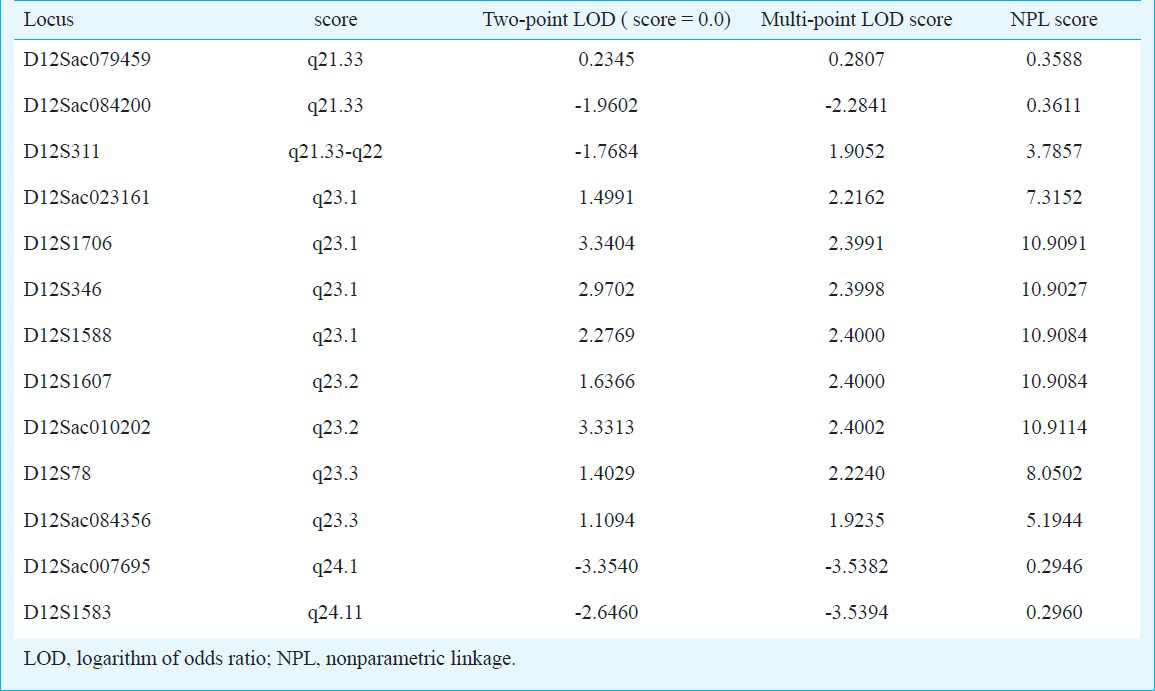
Linkage analysis suggested that an affected disorder is linked to a locus in the chromosome interval 12q23.1 to 12q23.3; two-point parametric analysis showed a logarithm of odds (LOD) score of 2.97 for the marker D12S346 (12q23.1) and 1.40 for the marker D12S78 (12q23.3). Fine mapping and haplotype analysis subsequently confirmed that eight continuous markers (D12Sac023161, D12S1706, D12S346, D12S1588, D12S1607, D12Sac010202, D12S78, D12Sac084356) had positive LOD with a maximum two-point LOD score of 3.34 for the marker D12S1706 and a maximum multi-point LOD score was 2.4002 for D12Sac010202, their NPL scores were 10.9091 for D12S1706 and 10.9114 for D12Sac010202.
Interpretation & conclusions:
A novel locus for essential hypertension was identified on chromosome 12q23.1 - q23.3. This finding implies that the region 12q23.1 to 12q23.3 might encompass a susceptible gene that caused hypertension in this Chinese family.
Keywords
Chromosome 12
haplotype analysis
hypertension
linkage
microsatellite
Essential hypertension (EH) has long been considered a complex disease impacted by genetic and environmental components. It is one of the most common cardiovascular diseases, and widely accepted as a significant risk factor for heart attack, stroke and kidney failure1. In recent years, more than ten candidate loci have been reported to contribute to the susceptibility of EH2, whereas having susceptibility gene remains unknown due to its genetic heterogeneity. Racial differences, genetic heterogeneity, and environmental factors hinder the genetic studies of EH2. Different susceptibility genes for hypertension are found to be involved in different cohorts with EH2. In the present study, we applied genome wide linkage analysis to localize a susceptibility locus in a single Chinese family with EH.
Material & Methods
An EH family from Shanghai, PR China was identified through a proband who had the typical features of hypertension. The proband (IV: 9 in Fig. 1) was a male born in 1956, who was diagnosed essential hypertension (BP ≥140/90 mmHg) at the age of 40 when he came to hospital in 1997. His blood pressure reached the highest of 150/110 mmHg. He was given amlodipine (5 mg/day) to control his blood pressure. Electrocardiogram (ECG), renal function, lipid profile, plasma and 24 h urine ion, including plasma creatinine, triglyceride, total cholesterol, high density lipoprotein (HDL), low density lipoprotein (LDL), as well as plasma and 24 h urine sodium, potassium, chloride, etc. were in the normal ranges.
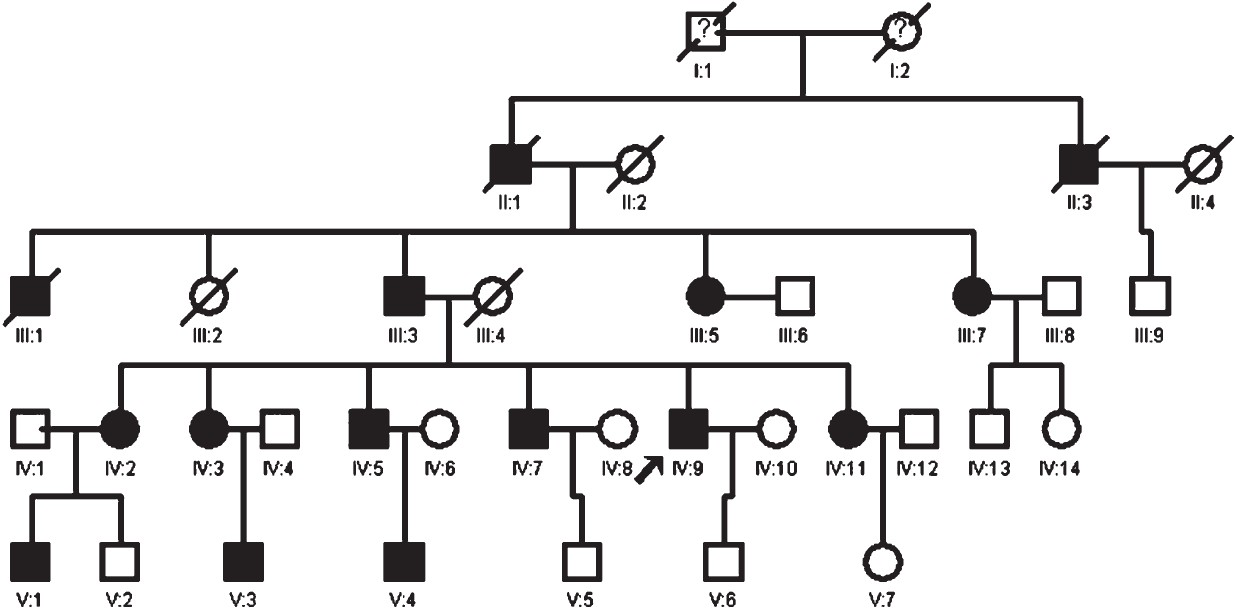
- Abridged pedigree of the EH-affected family.
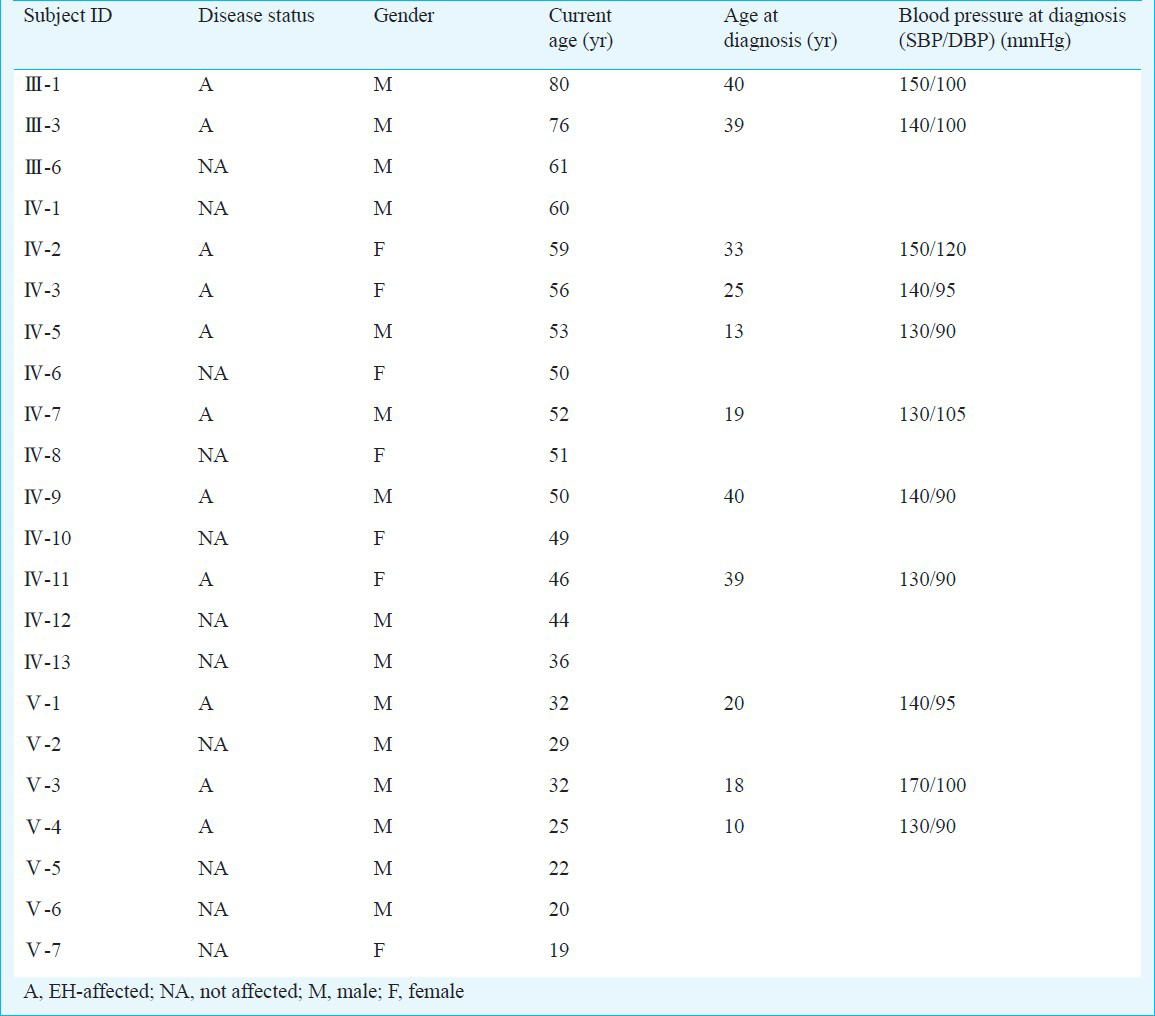
An individual was considered as hypertensive who had systolic blood pressure (SBP) ≥140 mmHg, or diastolic blood pressure (DBP) ≥90 mmHg, and was taking antihypertensive medicine for the treatment of hypertension. Normotensive controls were defined as individuals having a SBP ≤120 mmHg and DBP ≤85 mmHg and not taking any antihypertensives. Of the 22 family members of the proband, 11 were diagnosed with hypertension. In addition, early-onset of the hypertension was observed before the age of 40. Some of them were diagnosed as hypertensive earlier than age 20. Notably, they had high diastolic pressure, but high-normal systolic blood pressure, normal liver and kidney functions and lipid profile, plasma and 24 h urine sodium, potassium, chloride were all within normal range. Their clinical features are described in Table I.
Peripheral blood samples (2 ml) were obtained from these 22 subjects and genomic DNA was extracted from blood samples. Written informed consent from each subject was obtained before the study. The study protocol was approved by the Xinhua Hospital ethics committee, Shanghai Jiaotong University, School of Medicine, Shanghai.
Microsatellite markers: Genome-wide screen was performed with 424 polymorphic fluorescently labelled microsatellite markers, which were distributed over the 22 autosomes and the X chromosome. Among these, 418 markers were obtained from Perkin-Elmer Biosystem Division (ABI Prism Linkage Mapping Set version 2, Foster City, Calif., USA) which were packaged in a commercially available reaction kit. while the remaining six were custom-designed and custom-synthesized to fill the occasional oversized genotyping gaps.
Polymerase chain reaction (PCR) amplification: PCR reactions were carried out by using 5 ng of genomic DNA as a template in a mixture of 1 × PCR buffer, 15 mM MgCl2, 1 mM dNTP, 0.2 μM of each primer, and 0.2 U of Hotstar Taq polymerase (QIAGEN, Germany)in a final volume of 5 μl. The thermocycling conditions were 95°C for 15 min, followed by 15 cycles at 94°C for 30 sec, 63°C for 60 sec (decreasing 0.5°C per cycle for touch down), and 72°C for 90 sec, followed by 25 cycles at 94°C for 30 sec, 56°C for 60 sec, and 72°C for 90 sec, completed by a 10 min final extension step at 72°C. The amplified products were stored at 4°C until use.
Electrophoresis and genotype analysis: Fluorescent labelled amplification products were electrophoresed on an ABI PRISM® 3730 Genetic Analyzer with GeneScan 500HD ROX standard size label (Applied Biosystems, USA) and analyzed with Genescan 3.5 software (Applied Biosystems, USA). Alleles were analyzed with Genotyper 3.6 program (Applied Biosystems, USA).
Linkage and haplotype analyses: Two-point and multi-point parametric linkage analyses were performed by using the GENEHUNTER program (version 2.1, USA), assuming a genetic model featuring autosomal dominance a disease-allele frequency of 0.0001, evenly shared allele frequency, zero phenocopy rate, no sex difference, and full penetrance. Nonparametric linkage (NPL) analysis was calculated using the multipoint algorithm in GENEHUNTER. Haplotypes were reconstructed on the software program CYRILLIC version 2.1 (http://www.cyrillicsoftware.com/). The statistical power to detect linkage was evaluated by simulation using the computer program SIMLINK (http://csg.sph.umich.edu/boehnke/simlink.php)3.
Mutation screening on candidate genes: Based on the results of genome-wide linkage analysis, the main candidate genes in the region were analyzed, including SLC5A8, SLC17A8, SLC25A3, SLC41A2, IGF1, TRPV4, NR1H4, NR2C1, ELK3, SOCS2, LTA4H, NUDT4, TXNRD1, IKIP, RIC8B, ATP2B1. The coding and promoter regions, as well as their flanking intronic sequences of these genes were directly sequenced according to standard methodology on an ABI 3730 DNA automated sequencer (Applied Biosystems, USA).
Results
Linkage analysis: According to genome-wide parametric linkage analysis, the markers showing logarithm of the odds ratio (LOD) scores >1 were found on chromosome 12 in D12S346 (96 cm) and D12S78 (103 cm) loci, their two-point LODs were 2.9702 for D12S346 and 1.4029 for D12S78, and their multi-point LODs were 2.3998 for D12S346 and 2.2240 for D12S78, respectively. Similarly, in the NPL analysis, the NPL scores were 10.9027 for D12S346 and 8.0502 for D12S78 (Table II). Therefore, additional more microsatellite markers were subsequently used to confirm and fine map the locus in this region.
Eight continuous microsatellite markers analyzed (D12Sac023161, D12S1706, D12S346, D12S1588, D12S1607, D12, D12S78, and D12Sac084356) on chromosome 12q displayed positive LOD scores. Two-point and multi-point linkage analysis and NPL analysis all showed the susceptibility loci to EH in this family might be localized in q23.1-q23.3 of chromosome 12. The maximum two-point LOD score was obtained on D12S1706 (LOD = 3.34, θ = 0), and the maximum multi-point LOD score was 2.4002 on D12Sac010202, their NPL scores were 10.9091 for D12S1706 and 10.9114 for D12Sac010202, respectively (Table II).
Haplotype analysis: The haplotype analysis was conducted to estimate the most likely set of full typed marker alleles for each individual in the pedigree (Fig. 2). The disease-linked haplotype was found in individuals with EH (III:1, III:3, IV:1, IV:4, IV:5, IV:7, IV:9, IV:12, V:1, V:3 and V:4). This haplotype was localized between markers D12Sac023161 and D12Sac084356. Based on the human genome reference sequence (National Center for Biotechnology Information Build 36 version), this haplotype region is located on chromosome 12q23.1 - q23.3 (spanning approximately 14 Mbp). Haplotype analysis suggested that the region on chromosome 12q23.1- q23.3 might contain the disease gene for EH in this family.
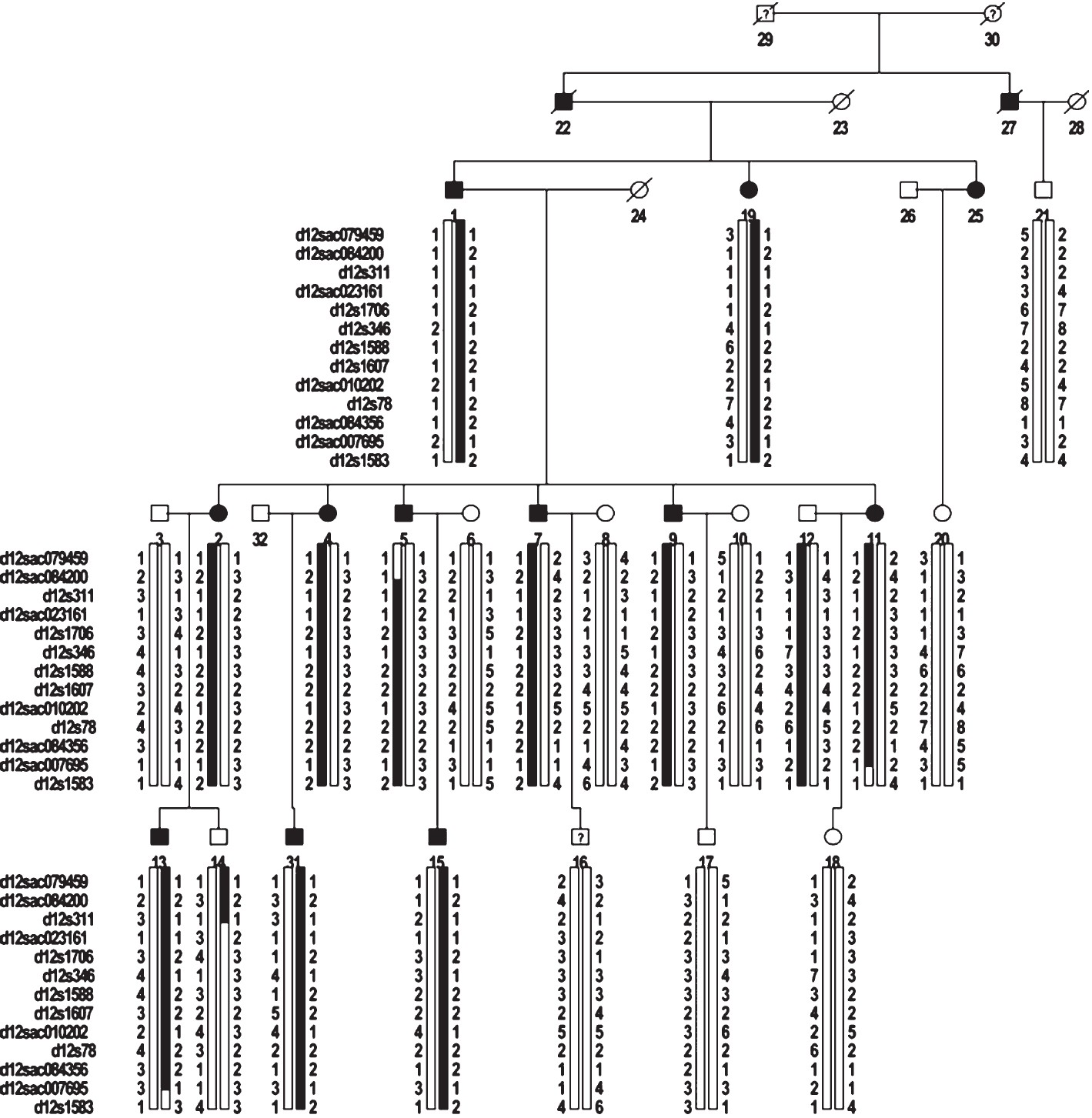
- Pedigree structure and haplotypes for the EH-affected family.
Mutation screening on candidate genes: The promoter, exon regions and their flanking intronic sequences of 16 candidate genes in or near this region, including SLC5A8, SLC17A8, SLC25A3, SLC41A2, IGF1, TRPV4, NR1H4, NR2C1, ELK3, SOCS2, LTA4H, NUDT4, TXNRD1, IKIP, RIC8B and ATP2B1, were sequenced by PCR. However, no mutations linkaged phenotype in this family, was detected. Additionally, the two SNPs, rs2681472 and rs2681492 in ATP2B1 gene, which were reported to have an association with hypertension in CHARGE Consortium4, were also genotyped. But the results showed that these two SNPs were not associated with hypertension phenotype in this family.
Discussion
With the rapid development of high throughput genotyping technologies, disease susceptibility loci and genes could be identified by genetic analysis. But the variation in clinical features of studies subjects and the environmental factors may have an effect on the participants, resulting in the poor repeatability5. Studying large kindred with disease-affected probands may reduce these interference factors and thus provide a useful method for identification of loci in various complex disorders.
In the present study, we identified a suspected locus for EH on chromosome 12q23.1- q23.3 in a Chinese family with inherited EH by genome-wide parametric linkage analysis and NPL analysis. EH has been recognized as a complex disease caused by genetic and environmental components, leading to the abnormal expression of some related genes and the enhanced level of blood pressure. A genome wide scan in Scandinavian sib-pairs demonstrated that 30-60 per cent of the variation in blood pressure was attributable to genetic factors, especially for patients with early onset primary hypertension6. Studies with families from Utah, U.S. revealed that individuals with two or more hypertensive first degree relatives developed hypertension approximately four times as frequent at onset by the age of 40, and gradually decreased with age. The predictive value from a family history of hypertension was negligible for hypertensive onset age of 705. These results suggest a critical role of genetic factors for disease development in patients with an early onset of hypertension.
Emerging evidences from genome-wide linkage studies have shown that susceptibility genes for EH are found in each human autosome except for chromosomes 14 and 222. Unfortunately, the genes responsible for human hypertension have not yet been extensively investigated due to the genetic heterogeneity, the differences in studied cohorts, environmental factors and the interaction between genetic and environmental components. Suggestion have been given to study the subgroups of EH-affected patients based on different clinical features. Such a study may provide a beneficial tool for exploring the disease mechanism. In this study, we investigated a five-generation Chinese pedigree comprising 11 EH-affected individuals. A high prevalence and early onset of hypertension was found in this family as revealed by clinical features. Most of the generation-IV relatives developed hypertension between age 20 and 40 yr, and the hypertension was developed in the generation-V individuals at the age of 20. In addition, their clinical features were very similar, as they had high diastolic blood pressure, but high-normal systolic blood pressure, implying that the genetic factors might contribute to the development of hypertension. Genome-wide scan and linkage analysis identified a principle locus for EH on chromosome 12q23.1-q23.3 (maximum two-point LOD score = 3.34 and multi-point LOD score = 2.4002), which partially overlaps the hypertension-related genetic region previously reported by Camp et al7. Multi-point linkage analysis confirmed that two regions localized to chromosome12q23.1-12q24.21 (LOD = 2.59) and chromosome 8p21.3-8p12 (LOD = 2.89) were closely associated with multiple blood pressure traits7.
The cohorts for Heart and Aging Research in Genome Epidemiology (CHARGE) Consortium was established to identify common genetic variation associated with complex traits4. Levy and his colleagues4 reported that there were several single nucleotide polymorphism (SNPs) loci in chromosome 12q21-12q24 with association to the levels of systolic and diastolic blood pressure and hypertension from 29,136 participants in the CHARGE Consortium4. SNP rs2681472 in ATP2B1 (on chromosome 12q21) had an association with diastolic blood pressure, while rs2681492 in ATP2B1 was correlated with systolic pressure4. Although these two SNPs showed no association with hypertension phenotype in our study, the findings in the CHARGE Consortium overlap with our results by linkage analysis4, suggesting that the susceptibility loci to human EH may be localized in q23.1-q23.3 of chromosome 12. ATP2B1, SLC41A2, IGF1, TRPV4, SOCS, and TXNRD1 genes distributed in this region are involved in regulating high blood pressure. PMCA1, encoded by the ATP2B1 gene, is a sarco (endo) plasmic reticulum Ca2+-ATPase, and acts as an essential regulator of free intracellular calcium8. SLC41A2 is an Mg2+ transporter involved in intracellular magnesium homeostasis in epithelial cells9. Insulin-like growth factor 1 (IGF-1) functions as an autocrine growth factor in a variety of cell types. Animal experiments showed that the adult IGF-1 mutant mice appeared to have high blood pressure10. Transient receptor potential vanilloid 4 (TRPV4), a mechanosensitive, swell-activated cation channel that is abundant in the renal distal tubules, acts as a flow sensor in flow-dependent K+ secretion11. The suppressors of cytokine signaling (SOCS) mediates Janus kinase and negatively regulates JAK/STAT signaling pathway12. Additionally, SOCS has been proposed to regulate IGF-1 signal pathway13. The selenoprotein thioredoxin reductase 1 (TrxR1), encoded by the TXNRD1 gene, is a key reductase in the oxidation-reduction system and has been demonstrated to guide actin polymerization in relation to cell membrane restructuring14. However, no mutations were found in the coding and promoter regions by sequencing analysis of these candidate genes, indicating that these genes may not be the disease-causing genes. We did not investigate the intron and intergenic regions of these candidate genes. The reported results showed that the intron region could regulate gene expression15, and the copy number variants of the gene were also related to protein expression through disrupting normal gene function, leading to the presence of abnormal phenotype1617. Therefore, we could not exclude the possibility that EH susceptibility loci might reside in the intron and intergenic regions of these candidate genes.
In conclusion, we identified a novel locus for EH on chromosome 12 q23.1- q23.3 in a Chinese family. However, no mutations were observed in the promoters and exons of 16 candidate genes in this region. Further research is needed to determine the causative gene(s) located on the chromosome 12q23.1- q23.3.
References
- A review of the genetics of essential hypertension. Curr Opin Cardiol. 2007;22:176-84.
- [Google Scholar]
- SIMLINK: A program for estimating the power of a proposed linkage study by computer simulation. In: Version 4.12. USA: University of Michigan; 1997.
- [Google Scholar]
- Genome-wide association study of blood pressure and hypertension. Nat Genet. 2009;41:677-87.
- [Google Scholar]
- Dissecting complex traits: recent advances in hypertension genomics. Genome Med. 2009;1:43.1-43.5.
- [Google Scholar]
- A genome wide scan for early onset primary hypertension in Scandinavians. Hum Mol Genet. 2003;12:2077-81.
- [Google Scholar]
- Genome-wide multipoint parametric linkage analysis of pulse pressure in large, extended utah pedigrees. Hypertension. 2003;42:322-8.
- [Google Scholar]
- Aortic smooth muscle and endothelial plasma membrane CA2+ pump isoforms are inhibited differently by the extracellular inhibitor caloxin 1b1. Am J Physiol. 2006;290:1341-9.
- [Google Scholar]
- Functional characterization of the mouse [corrected] solute carrier, SLC41A2. Biochem Biophys Res Commun. 2005;330:701-5.
- [Google Scholar]
- Elevated blood pressure and enhanced myocardial contractility in mice with severe IGF-1 deficiency. J Clin Invest. 1996;98:2648-55.
- [Google Scholar]
- TRPV4 channel participates in receptor-operated calcium entry and ciliary beat frequency regulation in mouse airway epithelial cells. Proc Nat Acad Sci USA. 2008;105:12611-6.
- [Google Scholar]
- Cloning and functional analysis of new members of STAT induced STAT inhibitor (SSI) family: SSI-2 and SSI-3. Biochem Biophys Res Commun. 1997;237:79-83.
- [Google Scholar]
- Interaction of human suppressor of cytokine signaling (SOCS)-2 with the insulin-like growth factor-I receptor. J Biol Chem. 1998;273:24095-101.
- [Google Scholar]
- Induction of cell membrane protrusions by the N-terminal glutaredoxin domain of a rare splice variant of human thioredoxin reductase 1. J Biol Chem. 2008;283:2814-21.
- [Google Scholar]
- Sequence diversity in CYP3A promoters and characterization of the genetic basis of polymorphic CYP3A5 expression. Nat Genet. 2001;27:383-91.
- [Google Scholar]
- Copy-number variation and association studies of human disease. Nat Genet. 2007;39(7 Suppl):S37-42.
- [Google Scholar]
- A study of CNVs as trait-associated polymorphisms and as expression quantitative trait loci. PLoS Genet. 2011;7:e1001292.
- [Google Scholar]






