Translate this page into:
BRCA1 promoter methylation & its immunohistochemical correlation in sporadic breast cancer
For correspondence: Dr Preeti Agarwal, Department of Pathology, King George’s Medical University, Lucknow 226 003, Uttar Pradesh, India e-mail: preavn@gmail.com
-
Received: ,
This article was originally published by Wolters Kluwer - Medknow and was migrated to Scientific Scholar after the change of Publisher.
Abstract
Background & objectives:
Studies have shown that apart from hereditary breast carcinomas, breast cancer susceptibility gene 1 (BRCA1) mutations conferring to its loss are seen in sporadic breast carcinomas (SBC) as well. The aim of the present study was to assess BRCA1 methylation in females presenting at King George’s Medical University, Lucknow, with SBC by both immunohistochemistry (IHC) and methylation PCR with respect to hormonal profile and various morphological prognostic parameters. The primary objective was to look for the association between BRCA1 protein expression and DNA promoter methylation.
Methods:
81 mastectomy specimens from SBC of invasive breast carcinoma (no special type) were included in this study. After a detailed morphological assessment, formalin fixed paraffin embedded tissue from a representative tumour area was selected for BRCA1 IHC by heat-mediated antigen retrieval under high pH and DNA extraction and further bisulphate treatment. BRCA1 was studied for methylation by methylated and unmethylated PCR-specific primers.
Results:
BRCA1 promoter methylation was present in 42/81 (51.9%) participants, with significant BRCA1 protein loss (72.7%; P=0.002). A significant association between BRCA1 loss and hormonal profile was found (P=0.001); maximum in triple negative breast carcinoma (TNBC) (72%; 18/25). Most of the TNBC also harboured methylation (68%). Although not significant grade II and III tumours, lymph vascular invasion, ductal carcinoma in situ, and nodal metastasis (≥3) were seen in a higher percentage in methylated tumours. Mortality in SBC was significantly associated with BRCA1 loss (30.3%; P=0.024).
Interpretation & conclusions:
Study results highlight the concept of “BRCAness” in SBC as well. Hence, we can confer that identification of BRCA1 loss in SBC can make it a perfect candidate for poly ADP-ribose polymerase inhibitors or cisplatin-based therapy like hereditary ones.
Keywords
BRCA1
Breast cancer susceptibility gene 1
immunohistochemistry
promoter methylation
sporadic breast carcinoma
triple negative breast cancer
Breast carcinoma accounts for 24.2 per cent of female carcinomas worldwide resulting in the highest mortality rate of about 16 per cent according to the Global Statistics for 2020 and 20401. The Cancer Genome Atlas (TCGA) Network breast carcinoma categorized it into four main subtypes according to various treatment modalities as Luminal A and B [oestrogen receptor/progesterone receptor (ER/PR) positive], Her2 (human epidermal growth factor receptor 2) positive with or without ER or PR expression (Her2neu enriched) and ER, PR, Her2 negative [triple-negative breast carcinoma (TNBC)]2.
Breast cancer susceptibility gene 1 (BRCA1) is a classic tumour suppressor gene located on chromosome 17 and is usually found in hereditary breast and ovarian carcinoma and associated with homologous recombination, DNA repair, and in transcription. BRCA1 mutation has been related to familial breast carcinoma only. Few literature from the Western world as well as from India have demonstrated the role of BRCA1 in sporadic breast carcinoma also3-5.
Familial breast carcinoma is quite rare compared to sporadic breast tumours. The demonstration of BRCA1 protein loss and BRCA1 promoter methylation in malignant cells compared to normal mammary epithelial cells provide a substantial role of BRCA1 in sporadic tumours as well. Mostly, there is a loss of BRCA1 nuclear protein, in around 19 per cent of sporadic breast carcinomas (SBC), which have both nuclear and cytoplasmic BRCA1 protein loss6.
The aberrant, BRCA1 promoter hypermethylation, cytosine phosphate guanine (CpG) islands in promoter regions at 5’ end of BRCA1 induces downregulation expression of the BRCA1 protein expression. It can be detected in sporadic breast carcinoma, particularly in TNBCs7.
BRCA1 deletion is usually associated with increased sensitivity to drugs that induce cross-links (platinum chemotherapy) and single- and double-stranded breaks (etoposide) in DNA7. The breaks in DNA are usually repaired by the repair pathway that involves base excision in which poly ADP-ribose polymerase 1 (PARP1) is one of the major components8. PARP inhibitors can be used as targeted therapy in BRCA1 loss cases.
Hence with the above knowledge, it is apparent that sporadic cancer may harbour BRCA1 loss, especially triple-negative cancers which have limited therapy options. BRCA1 loss will make such individuals candidates for platinum, etoposide-based therapy as well as PARP inhibitors8. Multiple research studies are being undertaken to clarify the role of BRCA1 expression in sporadic breast tumours, but there are many contradictory results among the various studies.
BRCA1 has been studied in sporadic breast cancers; both in the Western and Indian population7,9-11. The methodology used includes either molecular, mainly PCR or immunohistochemistry (IHC). Only a few studies have utilized both of these. BRCA1 genetic testing is time-consuming and expensive. In developing countries such as India, where molecular diagnostics are less accessible to many, this study intended to study BRCA1 using both molecular technique (methylation PCR) as well as IHC (protein expression) in formalin-fixed paraffin-embedded (FFPE) tissue samples to ascertain the association of BRCA1 alteration through both techniques. Moreover, studying expression by both techniques on the same tissue can clarify regarding the exact difference that molecular alteration can make at the protein level. Hence, the present work was undertaken to study the BRCA1 status in females presenting with sporadic breast cancer in a tertiary care centre in northern India by both IHC as well as molecular techniques.
The primary objective of this study was to assess the protein expression of BRCA1 in invasive ductal carcinoma of the breast – no special type (IDC-NST) by IHC in females with no familial history of breast cancer in first- and second-degree relatives. Furthermore, the BRCA1 status by both IHC and methylation PCR with respect to the hormonal profile of breast cancer, various morphological prognostic parameters, Nottingham grade and clinical outcome was assessed. The association of BRCA1 protein expression (IHC) and DNA promoter methylation was also assessed.
Material & Methods
The present study was conducted in the department of Pathology, King George’s Medical University, Lucknow, from June 2019 to September 2020 after approval from the Institutes Ethics Committee (ESR/262/Inst/UP/2013/RR-16). A written informed consent was obtained from all the study participants.
Inclusion & exclusion criteria: Cases of sporadic breast cancer (i.e. females with no history of breast cancer in first- and/or second degree relatives), in which mastectomy specimens were received and the histology diagnosis was IDC-NST were included in this study. These cases also had an adequate tumour mass and a written informed consent was obtained from the individual. Cases having an inadequate tumour sample, other subtypes of invasive breast carcinoma, but unwilling to participate were excluded from the study.
A total of 81 masectomy cases of IDC-NST were included in the study. An FFPE tissue block from the representative tumour area on gross examination of mastectomy specimens was selected for further IHC and methylation study. 3-4 μm thick sections on silane-coated slide from this block was used for IHC. All the paraffin blocks were stored at room temperature and when the molecular test was performed, ten sections (7-8 μm) were collected in Eppendorf vials for DNA extraction and methylation study.
Unremarkable breast parenchyma on gross examination which was 5 cm away from the primary tumour site and harbouring unremarkable terminal ductal units with no evidence of hyperplasia either typical or atypical on microscopy was taken as internal control for each case. Excised breast tissue received (n=5) on account of cosmetic breast reduction surgeries with unremarkable morphology and with fibroadenoma (n=5) were used for standardization and as external control (as mentioned in the data sheet of the primary antibody used).
Morphological examination: FFPE blocks were processed for routine haematoxylin and eosin staining. The histological types, ductal carcinoma in situ (DCIS), tumour size, lymphovascular invasion, necrosis, nodal status and margins were assessed.
Immunohistochemistry (IHC): A section showing normal ductal glands and the tumour was selected for IHC as described earlier9 using ER [Oestrogen receptor; Flex polyclonal rabbit -a Hu ER alpha, Clone EP1, ready to use (RTU); DAKO AS/AS+], PR (Progesterone; Flex Monoclonal Mo a Hu PR, Clone PgR636, RTU; DAKO AS/AS+), Her2neu (polyclonal rabbit a Hu c-erb2 oncoprotein, RTU ; DAKO AS/AS+) and BRCA1 [polyclonal rabbit, RTU (AR345-5R); BioGenex Laboratories, Fremont, USA].
Breast cancer susceptibility gene 1 (BRCA1) immunostaining and interpretation: The sections were deparaffinized using xylene and then rehydration was done through graded alcohol and distilled water. For antigen retrieval, sections were treated with citrate buffer in pressure cooker for 15 min at 120°C followed by cooling at room temperature. Peroxidase blocking was done using H2O2 (3%) for 10 min along with protein blocking for 10 min. Overnight incubation of slides with BRCA1 primary antibody at 4°C in moist chamber was done followed by thrice washing with TRIS [Tris (hydroxymethyl) aminomethane] buffer at pH 7.6. Slides were incubated with polymer for next 30 min and further incubated with secondary antibody for 30 min. Diaminobenzidine was used as chromogen.
For IHC interpretation, the stained sections were examined under ×400, and the expression of BRCA1 (both cytoplasmic and nuclear) was interpreted. No immunoexpression was considered negative or complete loss of BRCA1. Immunoreactivity scores of BRCA1 staining were calculated by adding the number representing the percentage of immunoreactive cells and the number representing staining intensity. Both nuclear and cytoplasmic expression was recorded separately and average of both scores was used as total. The total score of 0-3 was taken as loss of BRCA1 and 4-8 was considered expression12.
BRCA1 methylation: Genomic DNA extraction was done from FFPE tissue using commercially available DNA extraction kit (Purelink™ Genomic DNA Kit, Invitrogen, Thermo Fisher Scientific, USA). The extracted DNA was first put to bisulphate treatment. BRCA1 promoter methylation was studied using methylated and unmethylated PCR-specific primers. The primers were adapted from Esteller et al13, and Butcher and Rodenhiser14, and were synthesized by Integrated DNA Technology, USA. The primers annealed in the promoter region and flanked the transcription site of BRCA1.
The PCR mixture contained 10 pmol of each primer, 2.5 μl of 10X PCR buffer, 0.8 μl of 10 mM deoxynucleoside triphosphates (dNTPs) mix, 1U of Taq polymerase, 5 μl treated DNA, and nuclease-free water to adjust total volume of 25 μl. The amplifications were carried out with the following conditions: 95°C for 10 min; 35 cycles at 95°C for 15 sec 58°C for 45 sec; and 72°C for 45 sec. The product was run on electrophoresis and visualized on agarose gel after staining with ethidium bromide (Fig. 1).
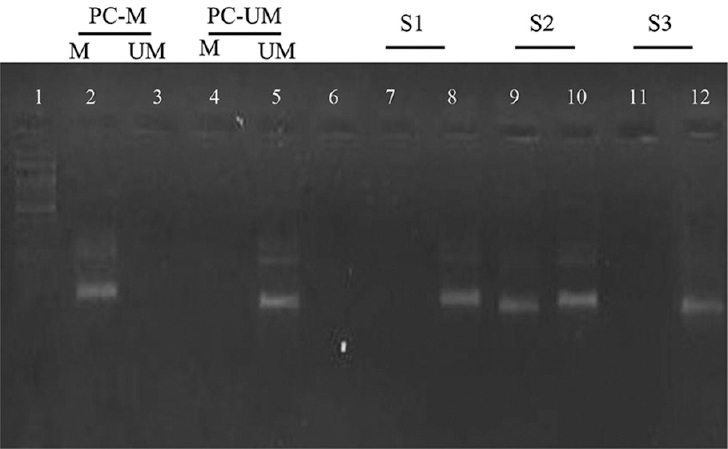
- Analysis by MSP PCR of the promoter region of BRCA1. Lane 1 indicates 100 bp ladder, Lane 2: positive control for methylated DNA; Lane 5: shows positive control for unmethylated DNA, Lane 6: negative control; Lane 7-8, 9-10 and 11-12 shows clinical samples with results for methylated and unmethylated PCR, respectively. The presence of a visible PCR product in those lanes marked UM indicates the presence of unmethylated genes of BRCA1; the presence of a product in those lanes marked M indicates the presence of methylated genes. MSP PCR, methylation-specific PCR; BRCA1, breast cancer susceptibility gene 1
Statistical analysis: Statistical Package for the Social Sciences for windows (SPSS version 21.0 Software; IBM Corp., Chicago, IL, USA) was used for statistical analysis.
Results
Clinicopathological parameters: The details of demographic and histomorphology variables studied are summarized in Table I. A total of 81 cases were categorized into four main hormonal subtypes i.e. luminal A (ER/PR+ Her2neu−; Ki67 <14%), luminal B (ER/PR+ Her2neu+; Ki67 >14%), Her2neu enriched (ER/PR–, Her2neu+) and TNBC (ER/PR/Her2neu all negative). Tumour with either one or more of the following morphological parameters were considered aggressive cancers:gross tumour size of >5 cm, tumour necrosis, presence of lymph, vascular invasion, presence of DCIS component, positive resection margins and involvement of ≥3 lymph nodes. Grade II tumours with tumour size between 2 and 5 cm and nodal metastasis were frequent in our study group.
| Characteristics | Number of cases, n (%) |
|---|---|
| Age (yr) | |
| ≤50 | 50 (61.7) |
| >50 | 31 (38.3) |
| Laterality | |
| Right | 33 (40.7) |
| Left | 48 (59.3) |
| Quadrant | |
| Upper outer | 35 (43.2) |
| Upper inner | 12 (14.8) |
| Lower outer | 9 (11.1) |
| Lower inner | 4 (4.9) |
| Central | 21 (25.9) |
| Size of tumour (cm) | |
| <2 | 3 (3.7) |
| 2-5 | 72 (88.9) |
| >5 | 6 (7.4) |
| Mean size of tumour (cm) | 3.63±1.22 (1-8) |
| Mitotic score | |
| Score 1 | 31 (38.3) |
| Score 2 | 39 (48.1) |
| Score 3 | 11 (13.6) |
| Mean mitotic count±SD (range) | 14.47±8.22 (2-40) |
| Nottingham grade | |
| Grade 1 | 17 (21) |
| Grade 2 | 50 (61.7) |
| Grade 3 | 14 (17.3) |
| Necrosis | |
| Absent | 19 (23.5) |
| Present | 62 (76.5) |
| Microcalcification | |
| Absent | 65 (80.2) |
| Present | 16 (19.8) |
| LVI | |
| Absent | 60 (74.1) |
| Present | 21 (25.9) |
| PNI | |
| Absent | 73 (90.1) |
| Present | 8 (9.9) |
| DCIS | |
| Absent | 43 (53.1) |
| Present | 38 (46.9) |
| Margins absent | 81 (100) |
| Lymph nodes metastasis | |
| Absent | 36 (44.4) |
| Present | 45 (55.5) |
| Number of lymph nodes involved | |
| <3 | 19 (42.2) |
| ≥3 | 26 (57.8) |
| Molecular subtyping (hormonal profile) | |
| ER/PR+ Her2neu−; (Luminal A) | 22 (27.2) |
| ER/PR+ Her2neu+; (Luminal B) | 10 (12.3) |
| ER/PR−, Her2neu+; (Her2neu enriched) | 24 (29.6) |
| ER/PR/Her2neu−; (TNBC) | 25 (30.9) |
| Outcome | |
| Succumbed to disease | 15 (18.5) |
| Alive | 66 (81.5) |
SD, standard deviation; LVI, lymphovascular invasion; PNI, perineural invasion; DCIS, ductal carcinoma in situ; ER, estrogen receptors; PR, progesterone receptors; TNBC, triple negative breast cancer
BRCA1 protein expression: The expression of BRCA1 in both internal and external controls was recorded. Staining in breast carcinoma cells was both nuclear and cytoplasmic and the intensity ranged from strong to absent (Fig. 2). BRCA1 was seen to be diffusely and strongly expressed by ducts of adjacent unremarkable breast parenchyma (Fig. 2A). BRCA1 protein expression was evaluated in all 81 cases, out of which complete loss of BRCA1 expression was seen in 33 (40.7%) cases and protein expression was present in 48 (59.3%) cases (Table II).
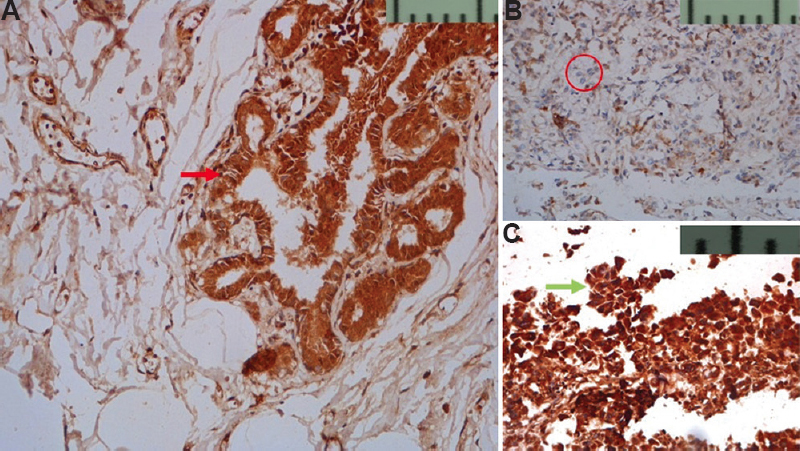
- (A) Nuclear and cytoplasmic expression of BRCA1 (red arrow) seen in normal TDLU (DAB, ×200). (B) Faint cytoplasmic expression in <25 per cent of tumour cells is seen, tumour cells are encircled as compared to normal TDLU in (A) (DAB, ×200). (C) Strong nuclear and cytoplasmic expression (green arrow) in >75 per cent of tumour cells as compared to normal TDLU in (A) (DAB, ×400) TDLU, terminal duct lobular unit
| BRCA1 IHC | Total (n=81) | |
|---|---|---|
| Nuclear expression | No expression | 25 |
| Expressed | 56 | |
| Cytoplasmic expression | No expression | 62 |
| Expressed | 19 | |
| Total BRCA expression | No expression | 33 |
| Expressed | 48 |
Comparison of BRCA1 protein expression and hormonal profile: The association between BRCA1 protein loss with hormonal profile was found to be significant (P=0.001). Least BRCA1 loss was seen in luminal A category 4/22 (18.2%) followed by Her2 enriched 7/24 (29.2%), luminal B 4/10 (40%). The maximum loss of protein expression was found in TNBC (72%; 18/25) (Table III). Both nuclear and cytoplasmic BRCA1 loss were seen in tumour cells. ER-negative tumours were found to be more frequent in BRCA1 protein loss category (62%).
| BRCA1 (IHC) | Total (n=81) | Luminal A (n=22), n (%) | Luminal B (n=10), n (%) | Her2neu enriched (n=24), n (%) | TNBC (n=25), n (%) |
|---|---|---|---|---|---|
| Loss | 33 | 4 (18.2) | 4 (40) | 7 (29.2) | 18 (72) |
| Expression | 48 | 18 (81.8) | 6 (60) | 17 (70.8) | 7 (28) |
χ2=16.090 (df=3); P=0.001.
BRCA1 protein expression and DNA promoter methylation: In our study, loss of BRCA1 protein was present in 33/81 cases, these 72.7%, (24/33) cases showed BRCA1 promoter methylation (Table IV). The association between DNA promoter methylation and BRCA1 protein loss was found to be significant (P=0.002) (Table IV).
| BRCA1 methylation status | n (%) | ||
|---|---|---|---|
| Unmethylated | 39 (48.1) | ||
| Methylated | 42 (51.9) | ||
| BRCA1 expression | Total (n=81) | Unmethylated (n=39), n (%) | Methylated (n=42), n (%) |
| No expression | 33 | 9 (27.3) | 24 (72.7) |
| Expressed | 48 | 30 (62.5) | 18 (37.5) |
χ2=9.720; P=0.002.
Correlation of BRCA1 promoter methylation with various prognostic parameters: Although no significant correlation was found between BRCA1 promoter methylation and various prognostic parameters including as necrosis, LVI, DCIS, tumour size >5 cm, and LN ≥3 positivity, LVI (26.2%), DCIS (47.6%) and LN ≥3 positivity (61.9%) were present in higher percentage in methylation cases compared to unmethylated tumours. High grade tumours (grade II and III) (54.7%) had promoter methylation, but this association was not found to be significant (P=0.322). When BRCA1 promoter methylation was studied with respect to hormonal profile, cancers in luminal A and luminal B categories showed methylation in eleven cases each. Three cases of Her2 were enriched and 17 cases of TNBC were methylated, respectively. There was a significant association (P=0.043) between BRCA1 promoter methylation and TNBC, most of the TNBC cases had methylation compared to non-TNBCs (68 vs. 32%) (Table V).
| BRCA1 methylation status | Total (n=81) | Luminal A (n=22), n (%) | Luminal B (n=10), n (%) | Her2 enriched (n=24), n (%) | TNBC (n=25), n (%) |
|---|---|---|---|---|---|
| Unmethylated | 39 | 9 (40.9) | 8 (80) | 14 (58.3) | 8 (32) |
| Methylated | 42 | 13 (59.1) | 2 (20) | 10 (41.7) | 17 (68) |
χ2=8.134 (df=3); P=0.043.
Association between BRCA1 protein expression and promoter methylation: Mortality in breast carcinoma cases was significantly associated (P=0.024) with BRCA1 protein loss (30.3%) compared to protein expression (10.4%). Promoter methylation showed more patients succumbed to disease compared to unmethylated cases (23.8 vs. 12.8%), however, this association was not found to be significant (P=0.203).
Discussion
BRCA1 is an important tumour suppressor gene which helps in maintaining genomic instability by uplifting homologous recombination repair. It is a DNA repair mechanism and in particular for double-stranded breaks13. BRCA1 is inherited in an autosomal dominant fashion and activated by p53 gene and other unknown mechanisms. Loss of heterozygosity of BRCA1 in sporadic breast carcinoma shares genotype as well as phenotype features with familial breast carcinoma with defect in the DNA repair pathway16.
Anomalous CpG hypermethylation in gene promoter region associated with loss of gene expression is an important mechanism for inactivation of tumour suppressor genes in malignant cells17. This is well reported in sporadic breast carcinoma, particularly in TNBC17. Women deficient in the BRCA1 gene are frequently found to have TNBC. TNBC is generally seen in younger age group, diagnosed in advanced stage with increased risk of metastasis and poor prognosis. These are not responsive to conventional receptor-target therapies also18.
Few SBC cases harbor BRCA1 phenotype with similar molecular characteristics like BRCA1/2 mutant breast carcinoma, known as “BRCAness” which makes it sensitive to PARP inhibitors as well as platinum-based therapy19.
BRCA1 loss has been associated with DNA promoter methylation and BRCA1 mutated cells are highly sensitized to inhibition of PARP enzyme19. PARP inhibitors cause replication fork stalling, leading to the formation of DNA substrates which further leads to the restart of replication by homologous recombination and are important for cell survival20. Hence, PARP inhibitors might be applicable in loss of BRCA1 function due to hypermethylation in sporadic tumours.
In the present study, total BRCA1 protein expression was assessed in 81 sporadic breast carcinoma cases and was found to be absent in 33 (40.7%) and expressed in 48 (59.2%). There is literature pertaining to nuclear, BRCA1 protein expression as well as cytoplasmic in breast tumour cells9. In the present study, nuclear as well as cytoplasmic expression was found in cases (Table II) and controls and none of the cases had only nuclear expression as observed by Al-Mulla et al21. Moreover, the expression of BRCA1 must always be interpreted with respect to adjoining unremarkable terminal ducts enclosed in any section from a tumour as is done for hormonal (ER/PR) markers, whenever possible.
Maximum BRCA1 loss in TNBC in our study suggests that similar hereditary breast carcinoma with BRCA1 mutation; sporadic cases may be frequently ER/PR/Her2neu negative too22. More ER-negative tumours harbored BRCA1 loss22 . This shows that loss of good prognostic markers such as ER also had BRCA1 loss, hence conferring poor prognosis to BRCA1 loss by its own virtue and its association with ER loss23. It has been suggested by Lee et al24 that loss of expression of nuclear BRCA1 (20.4%) positively correlated with a high histological grade while a complete loss of its nuclear expression correlated with other prognostic markers. Based on this the role of BRCA1 nuclear expression in the pathogenesis and prognosis of sporadic breast carcinoma was highlighted. The above findings (IHC results) also designate that high-grade tumours undergo some alteration at the molecular level. Studies have shown that BRCA1 loss in SBC could also be due to silencing due to promoter methylation9,25.
BRCA1 promoter methylation was present in 42/81 (51.9%) cases. Although no significance was found while assessing promoter methylation and various prognostic parameters in the present study; but the poor prognostic variables were more frequent in BRCA1 methylated tumours. Tian et al26 studied the concept of BRCAness in breast cancer by both BRCA1 germline mutations and methylation studies in TNBCs. They found that the BRCAness phenotype was largely associated with large tumour size (>2 cm), positive lymph node, tumour grade 3, high Ki67% levels (P=0.001), and basal like breast carcinoma. They concluded that BRCAness overall confers poor outcome to patients. Similar results were seen by Prajzendanc et al25 . In contrast almost equal distribution of grade, tumour size, and tumour necrosis in breast cancers in the present study could be the reason for non-significant results in the said context. The reason behind this observation could be that women in India seek medical attention only when the disease is quite advanced25,27. Furthermore, all hormonal profiles of breast cancers, with multiple prognostic variables were included in this study. Hence our results could be more representative of BRCA1 status and prognostic variables. Although multiple post-translational mechanisms also play a part in this, still some association of BRCA1 methylation in prognosis stands.
The correlation between BRCA1 protein loss and DNA promoter methylation was found to be significant (P=0.002; Table IV) similar to the findings of Bal et al9 and Miyamoto et al28. Hence, it can be deduced that BRCA1 promoter methylation was associated with decreased protein expression and epigenetic silencing of BRCA1 promoter regions.
We found significant correlation of molecular categories as compared to methylation results as seen when they were compared with protein expression (IHC). BRCA1 promoter methylation was found to be maximum in TNBC (Table V) similar to previous reports9.
However, the difference between the IHC and molecular results needs explanation. These may be explained by the following: First, molecular alterations could be either methylation silencing, deletion, point mutation, and others, and second, post-translation modifications.
Hence, reduced expression of BRCA1 protein and mRNA expression along with DNA promoter methylation in SBC cells compared with normal mammary epithelial cells provides a promising role of the BRCA1 gene in sporadic tumours. It is not only associated with a prognosis of these tumours but also provides evidence that patients harboring BRCA1 loss may benefit from PARP inhibitors (Olaparib) and platinum based chemotherapy.
The limitation of the present study is that we performed only methylation PCR in our study group to study the molecular alteration in SBC. India is a geographically and genetically variable country needs a larger multicentric study for BRCA1 evaluation in SBC to formulate its own BRCA1 testing and therapeutic guidelines.
To conclude, this study highlights the role of the BRCA1 gene in SBC among women in the Northern part of India. The loss of BRCA1 protein expression may be due to DNA promoter methylation in sporadic breast tumours as well apart from hereditary breast carcinomas which show BRCA1 mutations. TNBC tumours showed a significant loss of BRCA1, therefore highlighting the concept of BRCAness. These SBCs can therefore be benefited from PARP inhibitors and cisplatin based therapy as recommended in hereditary breast carcinomas. Hence, it is recommended that females with SBC, especially TNBC must be advised BRCA1 IHC scoring to identify cases which may benefit from the above chemotherapy.
Acknowledgment
Authors acknowledge histopathology laboratory staff of KGMU, Lucknow, for their technical support.
References
- Current and future burden of breast cancer: Global statistics for 2020 and 2040. The Breast. 2022;66:15-23.
- [Google Scholar]
- Germ-line BRCA1 mutations in women with sporadic breast cancer: Clinical correlations. J Clin Oncol. 1998;16:115-20.
- [Google Scholar]
- Evaluation of BRCA1, STAT-1 and STAT-3 expression in non familial breast cancer from North India: An interim analysis. J Clin Oncol. 2018;36(15 Suppl):e13010.
- [Google Scholar]
- Decreased expression of BRCA1 accelerates growth and is often present during sporadic breast cancer progression. Nat Genet. 1995;9:444-50.
- [Google Scholar]
- Epigenetic modifications in breast cancer and their role in personalized medicine. Am J Pathol. 2013;183:1052-63.
- [Google Scholar]
- The breast cancer susceptibility gene BRCA1 is required for subnuclear assembly of Rad51 and survival following treatment with the DNA cross-linking agent cisplatin. J Biol Chem. 2000;275:23899-903.
- [Google Scholar]
- BRCA1-methylated sporadic breast cancers are BRCA-like in showing a basal phenotype and absence of ER expression. Virchows Arch. 2012;461:305-12.
- [Google Scholar]
- Frequent alterations of homologous recombination repair pathway in primary and chemotolerant breast carcinomas: Clinical importance. Future Oncol. 2017;13:159-74.
- [Google Scholar]
- Differential association of BRCA1 and BRCA2 genes with some breast cancer-associated genes in early and late onset breast tumors. Ann Surg Oncol. 2004;11:1045-55.
- [Google Scholar]
- Estrogen receptor status by immunohistochemistry is superior to the ligand-binding assay for predicting response to adjuvant endocrine therapy in breast cancer. J Clin Oncol. 1999;17:1474-81.
- [Google Scholar]
- Promoter hypermethylation and BRCA1 inactivation in sporadic breast and ovarian tumors. J Natl Cancer Inst. 2000;92:564-9.
- [Google Scholar]
- Epigenetic inactivation of BRCA1 is associated with aberrant expression of CTCF and DNA methyltransferase (DNMT3B) in some sporadic breast tumours. Eur J Cancer. 2007;43:210-9.
- [Google Scholar]
- BRCA1/BARD1 E3 ubiquitin ligase can modify histones H2A and H2B in the nucleosome particle. J Biomol Struct Dyn. 2010;27:399-406.
- [Google Scholar]
- Sporadic breast carcinomas with somatic BRCA1 gene deletions share genotype/phenotype features with familial breast carcinomas. Anticancer Res. 2010;30:3445-9.
- [Google Scholar]
- Association of BRCA1 promoter methylation with rs11655505 (c.2265C>T) variants and decreased gene expression in sporadic breast cancer. Clin Transl Oncol. 2013;15:555-62.
- [Google Scholar]
- Evaluation of CD4+T-cells and CD8+T-cells in triple-negative invasive breast cancer. Indian J Pathol Microbiol. 2018;61:477-8.
- [Google Scholar]
- BRCA1 gene expression in breast cancer: A correlative study between real-time RT-PCR and immunohistochemistry. J Histochem Cytochem. 2005;53:621-9.
- [Google Scholar]
- Identification and use of biomarkers in treatment strategies for triple-negative breast cancer subtypes. J Pathol. 2014;232:142-50.
- [Google Scholar]
- Common origins of BRCA1 mutations in Canadian breast and ovarian cancer families. Nat Genet. 1994;8:392-8.
- [Google Scholar]
- Immunolocalization of BRCA1 protein in normal breast tissue and sporadic invasive ductal carcinomas: A correlation with other biological parameters. Histopathology. 1999;34:106-12.
- [Google Scholar]
- BRCA1 promoter methylation in peripheral blood is associated with the risk of triple-negative breast cancer. Int J Cancer. 2020;146:1293-8.
- [Google Scholar]
- Evaluation of the BRCAness phenotype and its correlations with clinicopathological features in triple-negative breast cancers. Hum Pathol. 2019;84:231-8.
- [Google Scholar]
- Delay in presentation to the hospital and factors affecting it in breast cancer patients attending tertiary care center in Central India. Indian J Cancer. 2015;52:102-5.
- [Google Scholar]
- Promoter hypermethylation and post-transcriptional mechanisms for reduced BRCA1 immunoreactivity in sporadic human breast cancers. Jpn J Clin Oncol. 2002;32:79-84.
- [Google Scholar]






