Translate this page into:
Molecular characterization & recombination analysis of complete enterovirus-88 isolated from acute flaccid paralysis cases in India
For correspondence: Dr Pragya D. Yadav, Scientist ‘E’ & Head, Maximum Containment Laboratory, ICMR-National Institute of Virology, Maximum Containment Facility, Sus Road, Pashan, Pune 411 021, Maharashtra, India e-mail: hellopragya22@gmail.com
-
Received: ,
This article was originally published by Wolters Kluwer - Medknow and was migrated to Scientific Scholar after the change of Publisher.
Abstract
Background & objectives:
Focus on non-polio enteroviruses (NPEVs) causing acute flaccid paralysis (AFP) due to myelitis has increased with the containment of the poliovirus. Enterovirus-B88 (EV-B88) has been associated with the AFP cases in Bangladesh, Ghana, South Africa, Thailand and India. In India, EV-B88 infection was linked to AFP a decade ago; however, to date, no complete genome has been made available. In this study, the complete genome sequence of EV-B88 was identified and reported from two different States (Bihar and Uttar Pradesh) in India using the next-generation sequencing technique.
Methods:
Virus isolation was performed on the three AFP suspected cases as per the WHO-recommended protocol. Samples showing cytopathic effects in the human Rhabdocarcinoma were labelled as NPEVs. Next-generation sequencing was performed on these NPEVs to identify the aetiological agent. The contiguous sequences (contigs) generated were identified, and reference-based mapping was performed.
Results:
EV-B88 sequences retrieved in our study were found to be 83 per cent similar to the EV-B88 isolate from Bangladesh in 2001 (strain: BAN01-10398; Accession number: AY843306.1). Recombination analyses of these samples demonstrate recombination events with sequences from echovirus-18 and echovirus-30.
Interpretation & conclusions:
Recombination events in the EV-B serotypes are known, and this work reconfirms the same for EV-B88 isolates also. This study is a step in increasing the awareness about EV-B88 in India and emphasizes future studies to be conducted in the identification of other types of EV present in India.
Keywords
Acute flaccid paralysis
Bihar
enterovirus-88
India
next-generation sequencing
rash
recombinant
Uttar Pradesh
Enterovirus (EV) genus comprises small, non-enveloped, positive-stranded RNA viruses belonging to the family Picornaviridae. The viral genome is approximately 7500 nucleotides long with a single open reading frame. It encodes for the four capsid proteins (VP1 to VP4) and the seven non-structural proteins1. The clinical manifestations associated with EV serotypes are acute haemorrhagic conjunctivitis, acute flaccid paralysis (AFP), arthritis, common cold, encephalitis, gastroenteritis, herpangina, meningitis, myocarditis, myelitis, pericarditis and upper and lower respiratory tract infections2-6. Some of the EV serotypes have found to be associated with diseases such as the hand, foot and mouth disease. Diagnoses based on clinical signs are often difficult for these serotypes, as many other viral aetiologies could cause the above-described symptoms.
Initially, EVs were classified depending on their pathogenesis in animals and humans, viz. coxsackie A virus, coxsackie B virus, poliovirus and echovirus1, and later, based on serotyping, these were classified into 64 different serotypes1. However, due to high genetic and antigenic diversity, even serotyping using the classical neutralization test became difficult to distinguish the members in this genus. Currently, a sequential number is given to enteroviruses, which are further grouped based on the phenotypic and genetic similarity. At present, enteroviruses have four subdivisions EV A-D7.
Enteroviruses (EV-B) consist of coxsackieviruses (CV-A9 and CV-B1 to 6), more than 20 EV-B serotypes and many echoviruses. The primary route of transmission of this species is by the faecal–oral route8. However, these are also found to be associated with mild respiratory infections, hand, foot and mouth disease, hepatitis, pleurodynia, herpangina and even severe infections causing myocardial infarction and meningitis9. There are studies also supporting their involvement in the onset of type I diabetes10. EV-B88 is a new addition to the EV-B species, which has been identified using molecular typing from stool samples collected from Bangladesh in an AFP surveillance during 200111. AFP surveillance carried out from 2007 to 2008, in Bangladesh, in school, children identified a few stool samples positive for EV-B8812. In India, the virus was detected through AFP surveillance systems in a few States (Kerala, Karnataka and Uttar Pradesh) between 2007 and 20096. After its initial detection, not much has been added regarding numbers detection of this particular serotype, except for a few sporadic viral detections in stool samples from India and other countries. To date, limited information about EV-B88 is available in the GenBank database, seven partial nucleotide sequences and one complete genome sequence (AY843306.1).
It is almost a decade how since awareness was gathered that surveillance for AFP leads to reporting of EV-B88 from India. However, to date, no complete genome of the virus has been made available or characterized from our country. This study reports the complete molecular characterization of the three EV-B88 serotypes detected from India [Bihar (2017) and Uttar Pradesh (2011)] through AFP surveillance along with its recombination analysis.
Material & Methods
This study was conducted at the National Polio Laboratory, Indian Council of Medical Research (ICMR)-National Institute of Virology (NIV)-Bangalore Unit, Bengaluru, Karnataka and BSL-4 Laboratoty, ICMR-NIV, Pune, Maharashtra, on AFP surveillance samples collected during 2011 and 2017. The study was approved by the Institutes Ethics Committee of ICMR-NIV. This study analyzed three AFP suspected cases, as per the case definition, from the AFP surveillance system conducted by the WHO-NPSP13. Two stool samples from each case were collected at an interval of 24 h and sent to the National Polio Laboratory at ICMR-NIV-Bangalore Unit, Bengaluru, under the cold chain.
Virus isolation: Virus isolation was performed as per the World Health Organization (WHO) recommended protocol14. Chloroform extraction was done on the stool samples and then inoculated into human rhabdocarcinoma (RD) and human poliovirus receptor-CD155 expressing recombinant L20B cell cultures and observed for any cytopathic effect (CPE). Samples with no changes in both cell cultures were labelled as negative. The samples showing CPE only in RD cell lines were labelled as NPEV. The tissue culture fluid of samples showing CPE in L20B cell lines was harvested and inactivated at 80°C and further identified by intratypic differentiation (ITD).
Molecular testing for virus identification: ITD is a screening tool where L20B-positive samples are subjected to real-time reverse transcriptase (rRT) PCR to identify EV and serotypes of polioviruses such as PV1, PV2 and PV3 and also non-Sabin-like PV isolates (primer-probe kits: courtesy CDC Atlanta). The isolates identified as non-polio enteroviruses (NPEVs) growing in L20B were sent for virus characterization to the ICMR-National Institute of Virology, Pune, India.
Next-generation sequencing of virus isolates: RNA extraction of the received samples was done using the modified protocol in combination with the TriPure reagent and QIAmp Viral RNA extraction kit (Qiagen, Germany) as per the manufacturer’s instructions. RNA thus extracted was quantified using the Qubit 2.0 Fluorometer (Invitrogen, USA). Host ribosomal depletion, fragmentation, library preparation and quantification were done as described earlier15. The quantified library was normalized and loaded on the Illumina MiniSeq NGS platform.
Paired-end reads generated from the Illumina MiniSeq NGS platform were loaded on the CLC Workbench software version 11.0.1 (CLC, Qiagen, Aarhus, Denmark). Contiguous sequences (contigs) were generated for the three different cases, using the de novo assembly programme of the software. The contigs generated were further analyzed using BLAST. Reference-based mapping was performed after the identification of the aetiological agent.
Phylogenetic analysis of enterovirus: Reference sequences of the EV-B were downloaded from the GenBank database. Sequence alignment was performed for viruses in MEGA software version 7.016. The maximum likelihood method, along with the general time-reversible model + Gamma+ I (GTR+G+I) substitution model, was used to generate the phylogenetic tree. The robustness of the tree was assessed using a bootstrap replication of 1000 cycles. Pair-wise percentage identity and differences were obtained using CLC Genomics Workbench version 11.0 (QIAGEN, Aarhus, Denmark). The phylogenetic tree was displayed using the FigTree v1.4.2.
Recombination analysis of enterovirus: A full exploratory study was performed using the RDP4 software to identify the potential recombination site in the EV-B8817. The protein-coding nucleotide sequences were compared to some EV-B strains. Methods used in the exploratory analysis were RDP, GENCOV, Bootscan, MaxChi, Chimaera, Siscan and 3seq. These methods are the individual algorithms that are incorporated in the RDP4 software, and each of these methods is capable of predicting recombination17. The sequence was considered a recombinant when more than three methods (algorithms) implemented in the RDP4 software detected a recombination event.
Further, the recombination analysis was performed using Simplot version 3.5.1 (http://sray.med.som.jhmi.edu/SCRoftware/SimPlot) and RDP4 software for the sequences that were predicted to be recombinant using the RDP4 software17. The window size of 450 nucleotides was used for a similarity plot. A neighbour-joining tree (Kimura 2-parameter model) using 1000 pseudo-replicates was used for running the Bootscan analysis.
Results
All three cases were male with a mean age of 1.1 yr. All three cases reported weakness in the leg. The first case from Bihar reported symptoms during November 2017 and the second and third cases from Utter Pradesh reported symptoms during December 2011 and July 2011, respectively (Table I).
| Case number | District | State | Age | Sex | Month of onset | Year | Paralysis limb | First sample VI | Second sample VI | Molecular test |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | Jumai | Bihar | One year | Male | November | 2017 | Lower limb - leg | Negative | Positive both RD and L20B | NPEV by PCR |
| 2 | Bareilly | Uttar Pradesh | One year four months | Male | December | 2011 | Lower limb - leg | Positive both RD and L20B | Positive both RD and L20B | NPEV by PCR |
| 3 | Bareilly | Uttar Pradesh | One year | Male | July | 2011 | Lower limb - leg | Positive both RD and L20B | Negative | NPEV by PCR |
VI, virus isolation; NPEV, non-polio enteroviruses; PCR, polymerase chain reaction; RD, rhabdomyosarcoma
Virus isolation results of all three cases demonstrated that the virus was grown in both RD and L20B cell lines (Table I). Furthermore, the real-time RT-PCR ITD results revealed that all three specimens were positive for EV but negative for pan PV and its serotypes, suggesting the isolate to be NPEV (Table I).
Analysis of the contigs for all the three cases, using BLAST, identified sequences to be similar to EV-B88. Once the causative agent was identified, reference mapping was performed using the GenBank reference sequence (KX595289). A complete genome sequence of ~7,370 nucleotides was obtained for all three EV-B88 strains using a reference sequence (AY843306). These sequences were deposited in the GenBank with accession numbers MH118025, MH144601 and MG982664. The details of the percentage of reads mapped, and the genome length recovered are given in Table II.
| Accession number | Relevant reads | Percentage of read coverage | Genome length recovered (bp) |
|---|---|---|---|
| MG982664 | 498579 | 39.58 | 7369 |
| MH144601 | 731029 | 95.04 | 7421 |
| MH118025 | 983743 | 96.62 | 7427 |
The retrieved genomes of viruses were compared with other EV-B reference genomes available in the GenBank database. Figure 1 depicts a phylogenetic tree for the VP1 gene of EV-B sequences retrieved in this study with other reference sequences. It was observed that the VP1 region of the Indian EV-B88 sequences forms a close cluster with that of Bangladesh (strain: BAN01-10398; Accession number: AY843306.1). The VP1 region of the EVs from India has 0.7-1 per cent amino acid differences in their similarity. The VP4 gene regions of the Indian EV sequences have 100 per cent amino acid similarity. The overall difference in the percentage amino acid similarity within the Indian EV sequences ranges from 0 to 1.6 per cent for different genes.
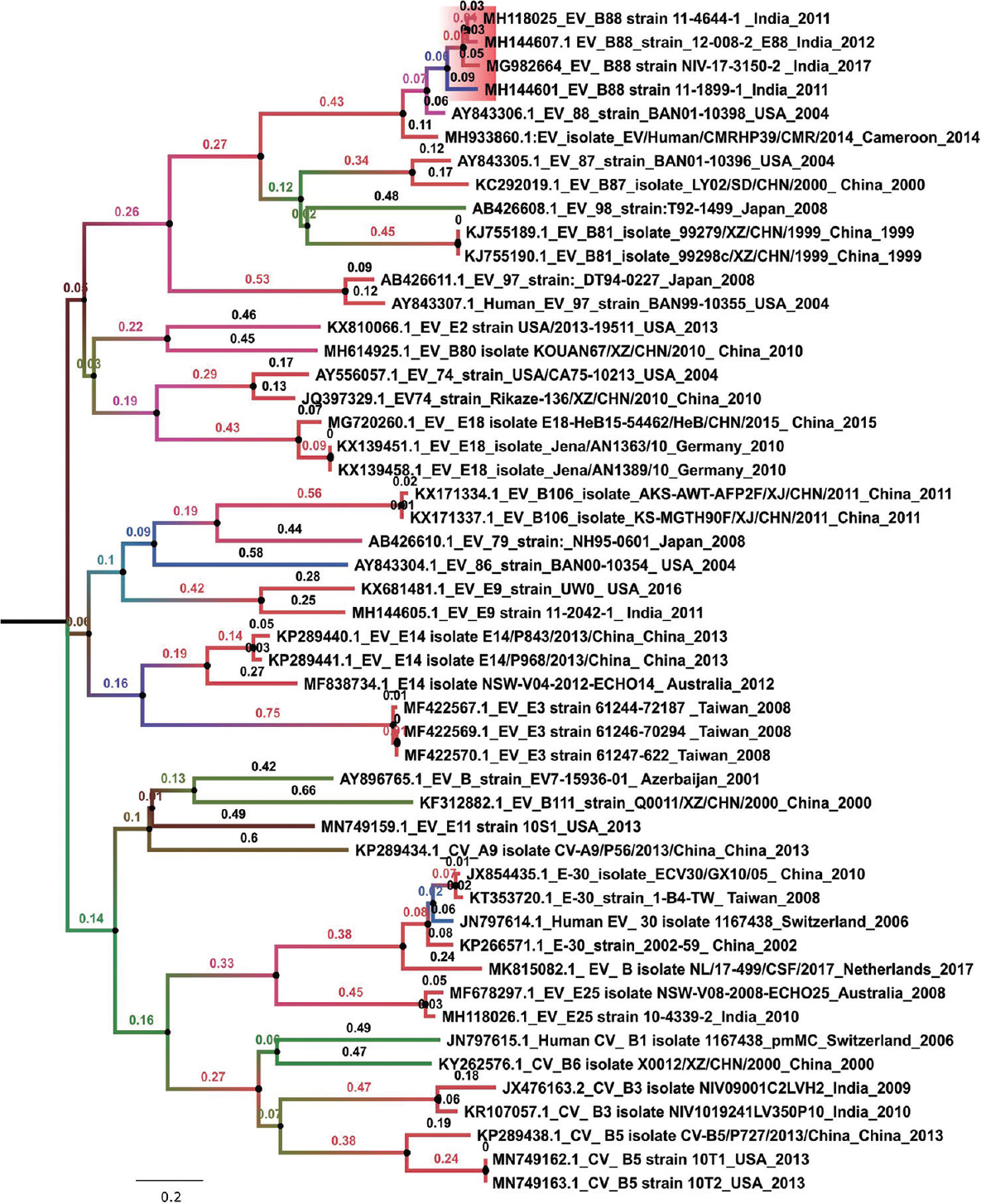
- Phylogenetic tree of EV-B88 VP1 gene isolated from Indian States (Bihar and Uttar Pradesh). Maximum likelihood method along general time reversible model + Gamma+ I was used for calculating the evolutionary distance among the EV-B88 (B) sequences. The robustness of the tree was evaluated using a bootstrap method involving 1000 re-samplings. The number displayed on the branches denotes the bootstrap value.
The VP1 region from the Indian EVs had a percentage amino acid similarity in the range of 96.6-98.3 to Bangladesh isolate. It was observed that the P1 region encoding for the non-structural genes (VP4-VP2-VP3-VP1) had an amino acid similarity in the range of 96.6-100 per cent. The P2 and the P3 regions encoding for the structural genes (2A-2B-2C) and (3A- 3B-3C-3D) respectively had a higher similarity to other EVs. Table III gives the percentage amino acid similarity of the EVs retrieved in this study and other EVs sequences downloaded from the GenBank with respect to Bangladesh (strain: BAN01-10398).
| EV-B88 accession number AY843306.1_EV_88_strain_BAN01-10398_USA_2004 | P1 region | P2 region | P3 region | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| VP1 | VP2 | VP3 | VP4 | 2a | 2b | 2c | 3a | 3b | 3c | 3d | |
| MH118025_EV_B88 strain 11-4644-1_India_2011 | 98.3 | 98.5 | 99.2 | 98.6 | 91.3 | 96 | 99.1 | 98.9 | 95.5 | 99.5 | 98.5 |
| MH144607.1 EV_B88_strain_12-008-2_E88_India_2012 | 98.3 | 98.9 | 98.7 | 100 | 92.6 | 98 | 98.2 | 98.9 | 95.5 | 97.8 | 97.8 |
| MH144601_EV_B88 strain 11-1899-1_India_2011 | 97.3 | 97.3 | 99.2 | 100 | 91.3 | 97 | 98.8 | 97.8 | 95.5 | 97.8 | 98.5 |
| MG982664_EV_B88 strain NIV-17-3150-2_India_2017 | 96.6 | 98.5 | 97.9 | 100 | 91.3 | 100 | 99.7 | 97.8 | 95.5 | 97.8 | 97.4 |
| MH933860.1:EV_isolate_EV/Human/CMRHP39/CMR/2014_Cameroon_2014 | 96.2 | 98.9 | 100 | 100 | 91.3 | 99 | 99.1 | 95.5 | 86.4 | 97.3 | 98.1 |
| AB426608.1_EV_98_strain: T92-1499_Japan_2008 | 76.1 | 86.2 | 87 | 82.6 | 91.3 | 94.9 | 98.5 | 100 | 90.9 | 98.9 | 97.8 |
| AY843305.1_EV_87_strain_BAN01-10396_USA_2004 | 73.6 | 83.5 | 84.5 | 76.8 | 91.9 | 98 | 98.2 | 96.6 | 100 | 98.9 | 98.9 |
| KJ755189.1_EV_B81_isolate_99279/XZ/CHN/1999_China_1999 | 73.6 | 81.6 | 82.4 | 72.5 | 89.9 | 97 | 97.9 | 96.6 | 95.5 | 98.4 | 98.1 |
| KJ755190.1_EV_B81_isolate_99298c/XZ/CHN/1999_China_1999 | 73.6 | 81.6 | 82.4 | 72.5 | 89.9 | 97 | 98.2 | 96.6 | 95.5 | 98.4 | 98.1 |
| KC292019.1_EV_B87_isolate_LY02/SD/CHN/2000_China_2000 | 72.9 | 83.5 | 84.9 | 78.3 | 91.9 | 100 | 98.8 | 95.5 | 100 | 97.8 | 98.5 |
| MG720260.1_EV_E18 isolate E18-HeB15-54462/HeB/CHN/2015_China_2015 | 70.3 | 80.8 | 77.8 | 75.4 | 89.3 | 94.9 | 97.3 | 100 | 95.5 | 97.3 | 98.1 |
| KX139451.1_Echovirus_E18_isolate_Jena/AN1363/10_Germany_2010 | 69.6 | 80.4 | 77.8 | 76.8 | 91.9 | 96 | 98.8 | 100 | 95.5 | 99.5 | 98.5 |
| KX139458.1_Echovirus_E18_isolate_Jena/AN1389/10_Germany_2010 | 69.2 | 80.4 | 78.2 | 76.8 | 91.3 | 96 | 98.8 | 100 | 95.5 | 98.9 | 98.5 |
| JQ397329.1_EV74_strain_Rikaze-136/XZ/CHN/2010_China_2010 | 67.4 | 79.6 | 75.7 | 73.9 | 89.3 | 97 | 98.8 | 100 | 95.5 | 98.4 | 97.6 |
| AB426611.1_EV_97_strain:_DT94-0227_Japan_2008 | 67 | 77 | 78.2 | 84.1 | 91.3 | 98 | 99.7 | 97.8 | 95.5 | 98.9 | 96.5 |
| AY556057.1_EV_74_strain_USA/CA75-10213_USA_2004 | 67 | 81.2 | 75.3 | 73.9 | 90.6 | 99 | 99.1 | 98.9 | 95.5 | 98.9 | 98.5 |
| AY843307.1_Human_EV_97_strain_BAN99-10355_USA_2004 | 67 | 77.8 | 77.8 | 82.6 | 91.9 | 97 | 98.2 | 100 | 95.5 | 99.5 | 98.1 |
| KP289440.1_EV_E14 isolate E14/P843/2013/China_China_2013 | 65.6 | 81.2 | 75.7 | 79.7 | 91.3 | 98 | 99.1 | 98.9 | 95.5 | 97.8 | 98.5 |
| KP289441.1_EV_E14 isolate E14/P968/2013/China_China_2013 | 65.6 | 77.7 | 75.3 | 78.3 | 91.3 | 99 | 98.8 | 96.6 | 90.9 | 97.8 | 98.3 |
| MF838734.1_E14 isolate NSW-V04-2012-ECHO14_Australia_2012 | 65.5 | 80 | 75.7 | 79.7 | 87.9 | 98 | 98.8 | 100 | 95.5 | 97.8 | 98.7 |
| KP266571.1_E-30_strain_2002-59_China_2002 | 64.7 | 76.2 | 71.8 | 82.6 | 90.6 | 100 | 98.8 | 98.9 | 95.5 | 96.7 | 98.5 |
| MH614925.1_EV_B80 isolate KOUAN67/XZ/CHN/2010_China_2010 | 64.6 | 80.4 | 75.7 | 79.7 | 92.6 | 98 | 98.2 | 98.9 | 90.9 | 97.8 | 98.5 |
| JN797614.1_Human echovirus 30 isolate 1167438_Switzerland_2006 | 64.3 | 78.2 | 71.8 | 81.2 | 90.6 | 98 | 98.5 | 97.8 | 90.9 | 98.9 | 98.5 |
| JX854435.1_E-30_isolate_ECV30/GX10/05_China_2010 | 64.3 | 76.2 | 71.8 | 82.6 | 89.9 | 97 | 99.1 | 100 | 90.9 | 97.8 | 99.4 |
| AY843304.1_EV_86_strain_BAN00-10354_USA_2004 | 64 | 77.3 | 74.9 | 81.2 | 90.6 | 97 | 99.4 | 100 | 95.5 | 97.8 | 98.3 |
| KX681481.1_Echovirus_E9_strain_UW0_USA_2016 | 64 | 81.5 | 77 | 84.1 | 91.9 | 94.9 | 98.5 | 100 | 95.5 | 97.3 | 97.8 |
| AY896765.1_EV_B_strain_EV7-15936-01_Azerbaijan_2001 | 64 | 77.4 | 77.7 | 81.2 | 87.9 | 96 | 99.4 | 100 | 95.5 | 98.4 | 98.1 |
| KT353720.1_E-30_strain_1-B4-TW_Taiwan_2008 | 64 | 76.6 | 71.4 | 82.6 | 89.9 | 97 | 99.4 | 100 | 90.9 | 98.9 | 99.1 |
| MH144605.1_Echovirus E9 strain 11-2042-1_India_2011 | 63.7 | 80 | 76.2 | 85.5 | 92.6 | 100 | 99.4 | 97.8 | 95.5 | 99.5 | 98.5 |
| KP289438.1_Coxsackievirus B5 isolate CV-B5/P727/2013/China_China_2013 | 63.7 | 77.8 | 71.8 | 84.1 | 91.3 | 98 | 98.8 | 97.8 | 90.9 | 98.4 | 98.3 |
| MK815082.1_Netherlands_2017 | 63.6 | 76.2 | 72.3 | 84.1 | 89.3 | 99 | 98.5 | 97.8 | 100 | 97.8 | 97.8 |
| KY262576.1_Coxsackievirus B6 isolate X0012/XZ/CHN/2000_China_2000 | 63.5 | 79.7 | 71.4 | 85.5 | 89.9 | 97 | 98.5 | 100 | 95.5 | 98.4 | 98.7 |
| KF312882.1_EV_B111_strain_Q0011/XZ/CHN/2000_China_2000 | 63.3 | 79.6 | 72.3 | 81.2 | 91.9 | 98 | 99.4 | 98.9 | 95.5 | 98.4 | 99.1 |
| MF422567.1_Echovirus E3 strain 61244-72187_Taiwan_2008 | 63.2 | 78.5 | 73.9 | 84.1 | 89.3 | 97 | 98.8 | 98.9 | 95.5 | 98.4 | 98.1 |
| MF422569.1_Echovirus E3 strain 61246-70294_Taiwan_2008 | 63.2 | 78.5 | 73.5 | 84.1 | 89.9 | 97 | 98.8 | 100 | 95.5 | 98.4 | 98.1 |
| MF422570.1_Echovirus E3 strain 61247-622_Taiwan_2008 | 63.2 | 78.5 | 73.5 | 84.1 | 89.9 | 97 | 98.8 | 100 | 95.5 | 98.4 | 97.8 |
| JN797615.1_Human coxsackievirus B1 isolate 1167438_pmMC_Switzerland_2006 | 63 | 78.5 | 74.8 | 81.2 | 90.6 | 94.9 | 97.9 | 97.8 | 90.9 | 98.9 | 98.5 |
| KR107057.1_Coxsackievirus B3 isolate NIV1019241LV350P10_India_2010 | 62.8 | 75.9 | 71.4 | 82.6 | 93.3 | 97 | 98.8 | 96.6 | 95.5 | 98.9 | 99.4 |
| KX810066.1_Echovirus E2 strain USA/2013-19511_USA_2013 | 62.7 | 78.5 | 75.3 | 75.4 | 89.3 | 96 | 99.4 | 97.8 | 90.9 | 98.4 | 98.9 |
| MN749162.1_Coxsackievirus B5 strain 10T1_USA_2013 | 62.5 | 79.3 | 73.5 | 82.6 | 91.2 | 94.9 | 99.7 | 97.8 | 90.9 | 97.8 | 98.5 |
| MN749163.1_Coxsackievirus B5 strain 10T2_USA_2013 | 62.5 | 79.3 | 73.5 | 82.6 | 91.2 | 94.9 | 99.7 | 97.8 | 90.9 | 97.8 | 98.7 |
| KP289434.1_Coxsackievirus A9 isolate CV-A9/P56/2013/China_China_2013 | 62.2 | 74.7 | 69.3 | 85.5 | 91.9 | 98 | 97.3 | 95.5 | 90.9 | 99.5 | 97.8 |
| KX171334.1_EV_B106_isolate_AKS-AWT-AFP2F/XJ/CHN/2011_China_2011 | 62.1 | 74.6 | 77 | 82.6 | 91.9 | 99 | 98.5 | 100 | 95.5 | 98.4 | 99.1 |
| KX171337.1_EV_B106_isolate_KS-MGTH90F/XJ/CHN/2011_China_2011 | 62.1 | 75 | 77 | 82.6 | 91.9 | 98 | 97.3 | 96.6 | 95.5 | 99.5 | 98.9 |
| JX476163.2_Coxsackievirus B3 isolate NIV09001C2LVH2_India_2009 | 62 | 76.2 | 71.8 | 81.2 | 91.3 | 99 | 99.1 | 98.9 | 95.5 | 98.4 | 98.7 |
| MN749159.1_Echovirus E11 strain 10S1_USA_2013 | 61.7 | 76.2 | 72.7 | 79.7 | 91.9 | 96 | 98.8 | 100 | 90.9 | 98.4 | 97.8 |
| MF678297.1_Echovirus E25 isolate NSW-V08-2008-ECHO25_Australia_2008 | 60.7 | 73.6 | 69.7 | 84.1 | 91.9 | 97 | 99.7 | 100 | 95.5 | 99.5 | 99.1 |
| MH118026.1_Echovirus E25 strain 10-4339-2_India_2010 | 60.7 | 72.8 | 69.7 | 85.5 | 91.9 | 98 | 98.8 | 98.9 | 95.5 | 98.9 | 98.3 |
| AB426610.1_EV_79_strain:_NH95-0601_Japan_2008 | 60.5 | 76.2 | 75.7 | 75.4 | 93.3 | 99 | 98.8 | 100 | 90.9 | 97.8 | 99.6 |
EV, enterovirus
The RDP4 analysis was performed to identify whether the EV-B88 sequences in this study were recombinant. Figure 2 depicts the similarity plot and Bootscan analysis of the MG982664 sequence using the Simplot software, respectively.
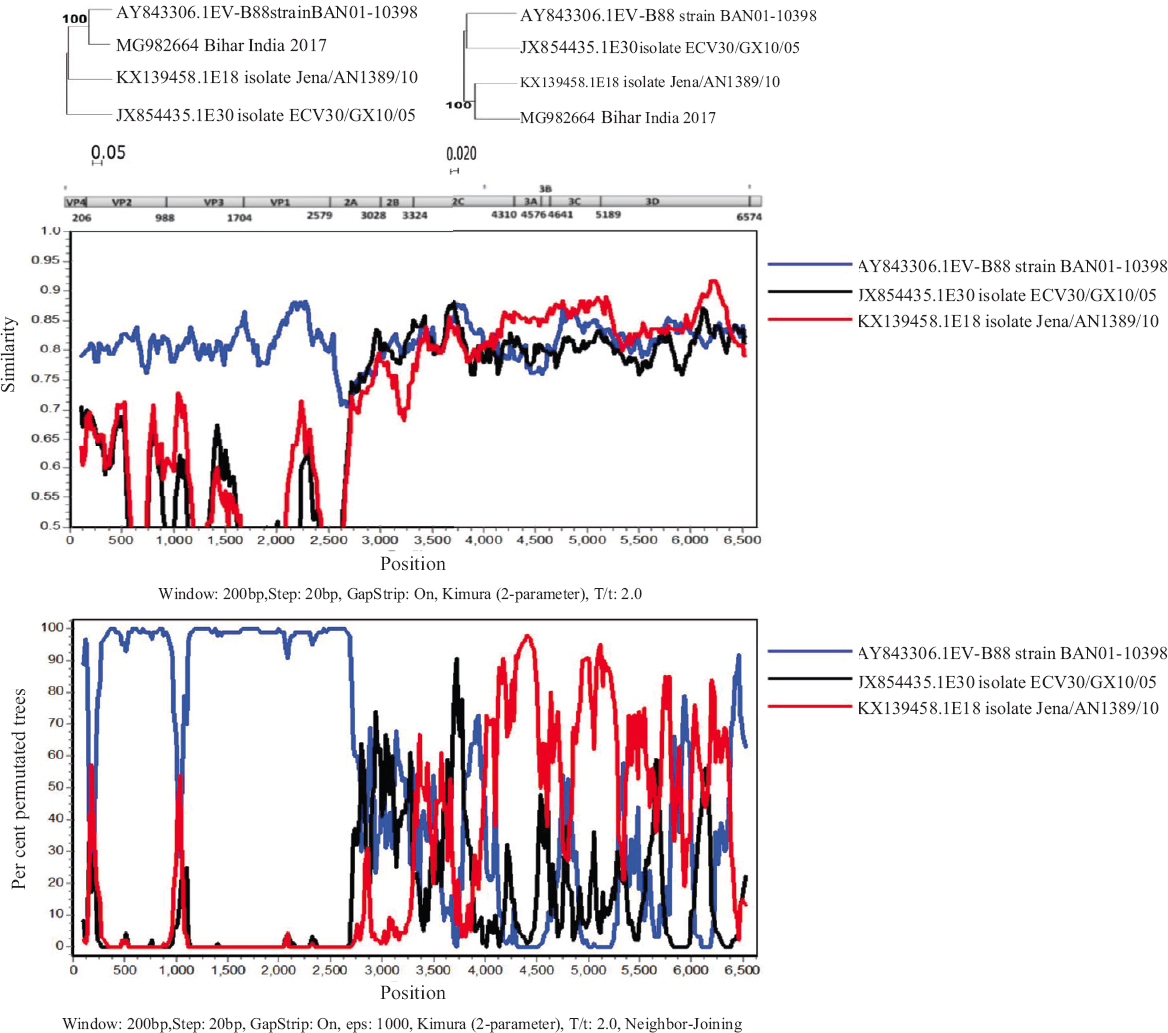
- Recombination analysis of MG982664 genome isolated from Bihar, India. Recombination analysis using Simplot and Bootscan using a window size of 200 nucleotides and a step size of 20. 1000 pseudo-replicates were used for running Bootscan analysis.
For understanding the reliability of the observed data, phylogenetic analysis was performed. Two separate phylogenetic trees were generated using the neighbour-joining method (K2P model) in MEGA software. A bootstrap replication of 1000 was used for statistical assessment. The clustering of the EV-B88 to different groups further confirmed that the retrieved sequences were recombinant.
Discussion
The present study identified three EV-B88 using the virus isolated from stool samples during an investigation carried out as part of the AFP surveillance system in UP and the southern part of Bihar State using NGS. Analysis of EV-B88 VP1 gene identified them to be closely related to Bangladesh (strain: BAN01-10398) of 2001. Further, it was also observed that the P2 and the P3 regions encoding for the structural genes had higher similarity to the other EVs as compared to Bangladesh (strain: BAN01-10398).
Sequence analysis is widely used to identify new EV species. This century has so far seen around 30 per cent in addition to EV genus. Other than Bangladesh and India, EV-B88 detection has also been reported from Ghana in association with enteric disease in children18, Western Cape province of South Africa in a rotavirus surveillance programme19 and Thailand (acute gastroenteritis in children) along with some other gastroenteritis virus with a prevalence of 1.5 per cent in the group affected20. Not much has been reported on EV-B88, except detections in association with AFP and gastroenteritis. Full-genome sequencing using Sanger’s method led to the retrieval of three different enteroviruses from the young children21. Nanopore direct RNA sequencing method was used to retrieve the EV sequences from the stool samples22.
This study analyzed the genomic sequences of EV-B88 from India and also looked upon the recombination within them. Each region of the EV-B88 performs a distinct role and hence can be individually evolved through mutation and recombination process. Lukashev23 found that the EV-B88 recombinant was most similar to the prototype in the VP4-VP2-VP3 region, demonstrating that the Indian human EV strain probably has an Bangladesh origin. Most of the recombination occurred near the structural region. The recently published article demonstrates the presence of diverse coxsackievirus A-10 (CV-A10) to be circulating in India24 that have been associated with AFP. A comparative analysis reveals that in India, CV-A10 infections are more common than EV-B88 infections that are linked to AFP24. The study is limited by its sample size and indicates the need for further sampling to concretize the finding.
Increasing awareness about the association of EVs with clinical symptoms indicates the need for further characterization of EVs that are isolated as part of a robust AFP surveillance in India. The group of cases that have isolated NPEVs in their stool samples and are presenting with some common clinical syndrome is worth further investigation. This study gives an idea of the probable pathogenic potential, indicating a need for further sampling to identify the burden of NPEV infection in the country.
Supplementary Fig. 1
Supplementary Fig. 1 Recombination analysis of MH144601 genome isolated from Uttar Pradesh, India: Recombination analysis using Simplot and Bootscan using a window size of 200 nucleotides and a step size of 20. 1000 pseudo-replicates were used for running Bootscan analysis.Supplementary Fig. 2
Supplementary Fig. 2 Recombination analysis of MH118025 genome isolated from Uttar Pradesh, India: Recombination analysis using Simplot and Bootscan using a window size of 200 nucleotides and a step size of 20. 1000 pseudo-replicates were used for running Bootscan analysis.Financial support & sponsorship: The study was funded by the World Health Organization (Reg No 2021/1185674) and ICMR-NIV, Pune.
Conflicts of Interest: None.
References
- Enteroviruses: Polioviruses, coxsackieviruses, echoviruses, and newer enteroviruses. In: Knipe DM, Howley PM, Griffin DE, Lamb RA, Martin MA, Roizman B, eds. Fields virology (5th ed). Philadelphia, PA: Lippincott Williams & Wilkins; 2007. p. :839-93.
- [Google Scholar]
- Genetic characterization of enterovirus strains identified in Hand, Foot and Mouth Disease (HFMD): Emergence of B1c, C1 subgenotypes, E2 sublineage of CVA16, EV71 and CVA6 strains in India. Infect Genet Evol. 2017;54:192-9.
- [Google Scholar]
- Molecular epidemiological study of enteroviruses associated with encephalitis in children from India. J Clin Microbiol. 2012;50:3509-12.
- [Google Scholar]
- The Coe virus: An apparently new virus recovered from patients with mild respiratory disease. Am J Hyg. 1958;68:272-87.
- [Google Scholar]
- Molecular surveillance of non-polio enterovirus infections in patients with acute gastroenteritis in Western India 2004-2009. J Med Virol. 2015;87:154-61.
- [Google Scholar]
- Antigenic diversity of enteroviruses associated with nonpolio acute flaccid paralysis, India 2007-2009. Emerg Infect Dis. 2012;18:1833-40.
- [Google Scholar]
- Enteroviruses in the early 21st century: New manifestations and challenges. Curr Opin Pediatr. 2016;28:107-13.
- [Google Scholar]
- Picornavirus and enterovirus diversity with associated human diseases. Infect Genet Evol. 2013;14:282-93.
- [Google Scholar]
- Enteroviruses in the pathogenesis of type 1 diabetes. Semin Immunopathol. 2011;33:45-55.
- [Google Scholar]
- Molecular identification of 13 new enterovirus types, EV79-88, EV97, and EV100-101, members of the species Human Enterovirus B. Virus Res. 2007;128:34-42.
- [Google Scholar]
- Characterizing the picornavirus landscape among synanthropic nonhuman primates in Bangladesh 2007 to 2008. J Virol. 2013;87:558-71.
- [Google Scholar]
- Polio laboratory manual. (4th ed). Available from: http://apps.who.int/iris/handle/10665/68762
- Complete genome sequencing of Kaisodi virus isolated from ticks in India belonging to Phlebovirus genus, family Phenuiviridae. Ticks Tick Borne Dis. 2019;10:23-33.
- [Google Scholar]
- MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 2016;33:1870-4.
- [Google Scholar]
- RDP4: Detection and analysis of recombination patterns in virus genomes. Virus Evol. 2015;1:vev003.
- [Google Scholar]
- Molecular characterization of enteric viral agents from children in northern region of Ghana. J Med Virol. 2008;80:1790-8.
- [Google Scholar]
- 2016. Available from: http://wiredspace.wits.ac.za/handle/10539/22385
- Multiple enterovirus genotypes circulating in children hospitalized with acute gastroenteritis in Thailand. Infect Genet Evol. 2017;55:324-31.
- [Google Scholar]
- Molecular characterization of three enteroviral strains isolated in Kuwait from young children with serious conditions. J Infect Dev Ctries. 2017;11:626-39.
- [Google Scholar]
- Whole-genome sequencing of human enteroviruses from clinical samples by nanopore direct RNA sequencing. Viruses. 2020;12:841.
- [Google Scholar]
- Role of recombination in evolution of enteroviruses. Rev Med Virol. 2005;15:157-67.
- [Google Scholar]
- Molecular diversity of Coxsackievirus A10 circulating in the southern and northern region of India [2009-17] Infect Genet Evol. 2018;66:101-10.
- [Google Scholar]






