Translate this page into:
Spectrum of GJB2 gene variants in Indian children with non-syndromic hearing loss
*For correspondence: madhulikakabra@hotmail.com
-
Received: ,
This is an open access journal, and articles are distributed under the terms of the Creative Commons Attribution-NonCommercial-ShareAlike 4.0 License, which allows others to remix, tweak, and build upon the work non-commercially, as long as appropriate credit is given and the new creations are licensed under the identical terms.
This article was originally published by Medknow Publications & Media Pvt Ltd and was migrated to Scientific Scholar after the change of Publisher.
Sir,
Hearing loss occurs in 155 out of every 100,000 births1, and in India it was reported as 291 per 100,000, according to the National Sample Survey, 20022. Non-syndromic hearing loss accounts for 70 per cent of all types of hereditary hearing loss. It can be autosomal recessive (80-85%), autosomal dominant (10-15%), X linked (1%) or mitochondrial (<1%). More than 130 loci and about 95 genes have been identified in non-syndromic hearing loss. Mutations in the GJB2 (DFNB1) gene are major contributors of pre-lingual hearing loss in various populations including India34. This locus was first mapped to chromosome 13q11 by linkage analysis in two large consanguineous Tunisian families with pre-lingual profound hearing loss5. GJB2 [connexin 26 (Cx26)] gene has two exons, of which only exon 2 (681 bp) is coding. Product of this gene, Cx26, forms a gap junction protein with four transmembrane domains6. GJB6 (Cx30), another gene of the connexin family, is also involved in the manifestation of non-syndromic hearing loss. Though many mutations of GJB2 have been reported in different studies, p. Trp24Ter mutation is known to be the most common cause in India378. Most studies reported a frequency of Cx26 mutations in Indian patients with non-syndromic hearing loss in a small sample. In this study, we report the spectrum of GJB2 variants as well as a novel variant in a large Indian cohort of 316 families with non-syndromic hearing loss.
These 316 families were enrolled between 2008 and 2015. Of these 70 families were enrolled from various States of India, including Gujarat (22), Delhi (22), Uttar Pradesh (UP, 21) and Madhya Pradesh (MP, 5) through the visit to various hearing impaired schools of the States. Of these 70 families, 28 were consanguineous and 42 non-consanguineous and had two or more hearing impaired children and at least one normal child. The remaining 246 families were referred to the Genetic Clinic, department of Pediatrics, All India Institute of Medical Sciences, New Delhi, India, for mutation analysis of GJB2 gene. In these 246 families, 37 families were consanguineous and some families had two or more hearing impaired children. The age group ranged from one to 18 years. In a total of 316 families, 537 affected individuals (292 males, 245 females) were screened. No formal sample size calculation was done for this study. Post hoc sample size was calculated (http://clincalc.com/stats/power.aspx) based on the frequency of GJB2 mutations (10%) in north India3 with allowable precision at three per cent. This gave a number of 400 individuals. Only cases of non-syndromic hearing loss were selected for the study. Families with recognizable syndromic/environmental causes of hearing loss were excluded. The study was approved by the institutional ethics committee and written consent was obtained from parents/ guardians of all participants even if the families were identified through schools. Permission to visit schools was taken from the concerned ministries.
Peripheral blood sample (2 ml) was taken from all the available affected members, and DNA was extracted by salting out method9. Coding exon 2 of GJB2 gene was amplified at annealing temperature (60°C) by primer set Cx26-2F and Cx26-2R. Polymerase chain reaction products were purified using enzyme (ExoI and SAP) to remove unincorporated deoxynucleotide triphosphates and primers. Bidirectional DNA sequencing of coding exon 2 was performed by four different primers Cx26-2F, Cx26-2R, Cx26-2FI and Cx26-2RI (Table I), to cover complete coding and splice site. Primers were designed by Primer3 software (http://primer3.ut.ee/). Sequence analysis was done with the help of Chromas Pro software (http://technelysium.com.au/wp/chromaspro), and sequences were aligned by MEGA (https://www.megasoftware.net) to a reference sequence (ENST00000382848) of GJB2 gene to identify sequence variants.

A total of 17 different variants (Table II) were identified in the coding exon 2 of GJB2 gene. Of these variants, 16 variants (c.35delG, c.71G>A, c.148G>A, c.223C>T, c.231G>A, c.238C>T, c.283G>A, c.313_326del14, c.340G>A, c.341A>G, c.370C>T, c.407dupA, c.439G>A, c.79G>A, c.380G>A and c.457G>A) are reported variants and c.616A>C is novel in nature. Of the 17 variants identified, 14 variants (13 reported mutations and one novel variant) were pathogenic variants and three variants were reported as polymorphisms.
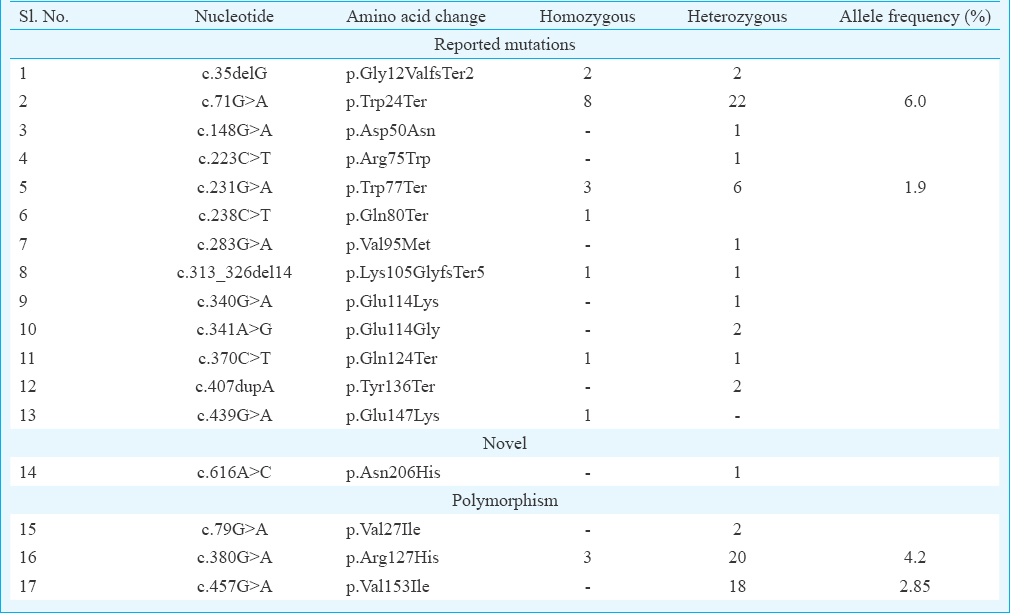
In the present study, a total of 27 families (20 homozygous and 7 compounds heterozygous) were found positive (8.5%) for Cx26 mutation. One novel variation c.616A>C (p. Asn206His) was also detected (Figure). The novel transversion c.616A>C was found in compound heterozygous state with c.71G>A (p. Trp24Ter) in a singleton non-consanguineous family. This change was predicted to be ‘probably damaging’ by PolyPhen2 (http://genetics.bwh.harvard.edu/pph2) and ‘damaging’ by SIFT (http://sift.jcvi.org) tools. According to amino acid conservation analysis of GJB2, it appears to be conserved across 52 homologous sequences (UniProtKB/Swiss-Prot, https://www.uniprot.org) and hence may be presumed to be pathogenic. In the study, p. Trp24Ter mutation was found to be the most common mutation and present in both homozygous and heterozygous state with an allele frequency of six per cent followed by p. Trp77Ter at allele frequency of two per cent. Two reported polymorphisms p. Arg127His (allele frequency 4.2%) and p. Val153Ile (allele frequency 2.85%) were also found in high frequency. Frequency of other reported mutations was very low (Table II).
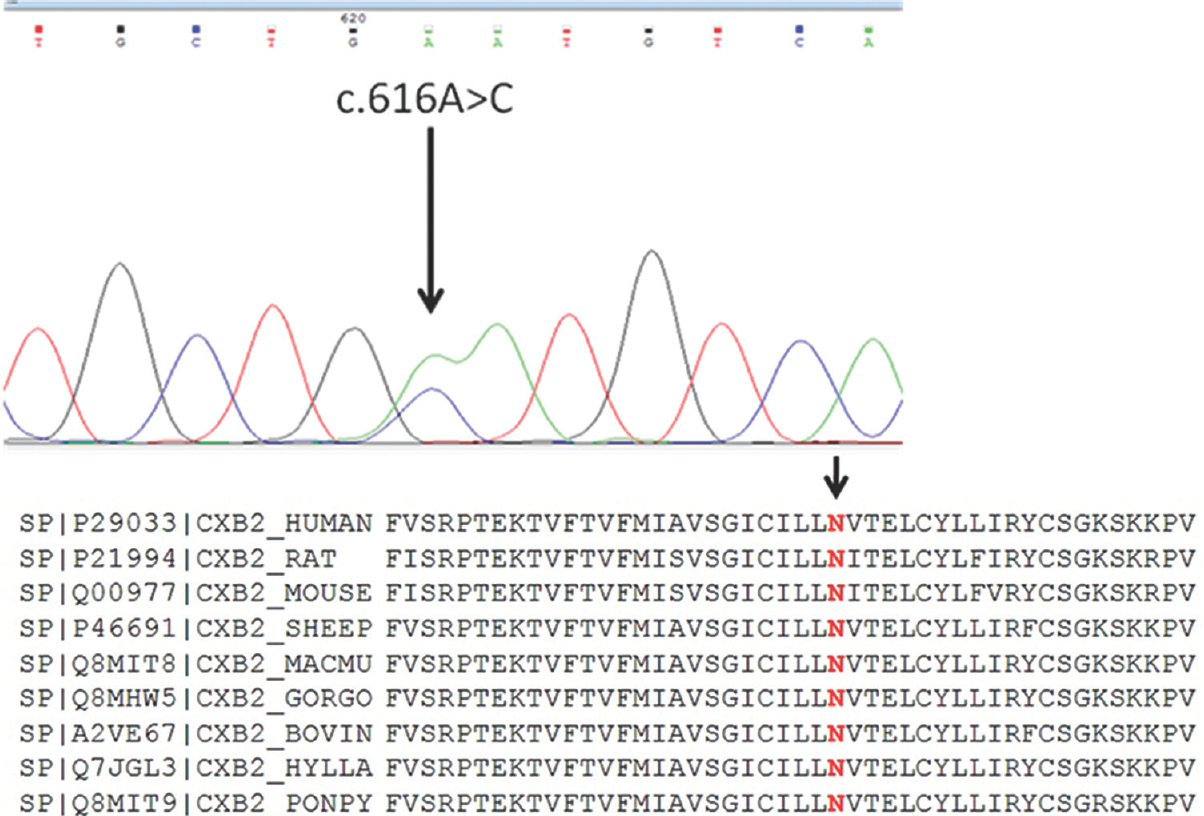
- Electropherogram showing heterozygous c.616A>C (p.Asn206His) change and protein sequence alignment showing conservation of amino acid asparagine at position 206 across the species.
Frequency of GJB2 mutations among hearing impaired individuals in India is much lower than European and North American population. The frequency also differs in south and north India3. This could be due to the prevalence of assortative mating in European and North American populations8. In Pakistan, frequency of Cx26 mutations was reported to be low in one study although consanguinity is twice as high as in India10. p. Trp24Ter mutation has been reported to be the most common cause for non-syndromic recessive hearing loss in India, followed by p. Trp77Ter, c.35delG and p. Gln124Ter3. p. Trp24Ter was detected as the founder mutation in India with a frequency of 33.9 per cent in a study done in Kerala11. Frequency of p. Trp24Ter was six per cent in our study predominately in north Indians. Apart from p. Trp24Ter and p. Trp77Ter, we found a truncating mutation p. Tyr136Ter which is due to duplication of adenine at nucleotide number 407. This mutation has also been reported in Indian population4. No regional variation of mutation spectrum of GJB2 gene was found in the study.
We also screened reported 342 kb deletion12 for GJB6 gene mutations in 70 families enrolled from UP, MP, Gujarat and Delhi through deaf schools and also sequenced the coding region in them but did not find any point mutation or the 342 kb deletion. Our results were concordant with earlier reported studies1314.
In conclusion, this study showed the spectrum of the GJB2 (Cx26) gene variants in children with non-syndromic hearing loss from northern (Delhi, UP) and western (Gujarat) regions of India. p.Trp24Ter was found to be the most common mutation noted in the GJB2 (Cx26) gene in patients with non-syndromic hereditary hearing loss. In addition, one novel change in compound heterozygous form was also found.
Financial support & sponsorship: Authors acknowledge the Department of Biotechnology, New Delhi, for financial support.
Conflicts of Interest: None.
References
- The Colorado newborn hearing screening project, 1992-1999: On the threshold of effective population-based universal newborn hearing screening. Pediatrics. 2002;109:E7.
- [Google Scholar]
- Ministry of Statistics and Programme Implementation, Government of India. Available from: http://mospi.nic.in/NSSOa
- Screening of families with autosomal recessive non-syndromic hearing impairment (ARNSHI) for mutations in GJB2 gene: Indian scenario. Am J Med Genet A. 2003;120A:180-4.
- [Google Scholar]
- GJB2-associated hearing loss: Systematic review of worldwide prevalence, genotype, and auditory phenotype. Laryngoscope. 2014;124:E34-53.
- [Google Scholar]
- A non-syndrome form of neurosensory, recessive deafness maps to the pericentromeric region of chromosome 13q. Nat Genet. 1994;6:24-8.
- [Google Scholar]
- High incidence of GJB2 gene mutations among assortatively mating hearing impaired families in Kerala: Future implications. J Genet. 2014;93:207-13.
- [Google Scholar]
- Spectrum and frequency of GJB2, GJB6 and SLC26A4 gene mutations among nonsyndromic hearing loss patients in eastern part of India. Gene. 2015;573:239-45.
- [Google Scholar]
- Functional consequences of novel connexin 26 mutations associated with hereditary hearing loss. Eur J Hum Genet. 2009;17:502-9.
- [Google Scholar]
- A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res. 1988;16:1215.
- [Google Scholar]
- Low prevalence of connexin 26 (GJB2) variants in Pakistani families with autosomal recessive non-syndromic hearing impairment. Clin Genet. 2005;67:61-8.
- [Google Scholar]
- High frequency of connexin26 (GJB2) mutations associated with nonsyndromic hearing loss in the population of Kerala, India. Int J Pediatr Otorhinolaryngol. 2009;73:437-43.
- [Google Scholar]
- A deletion involving the connexin 30 gene in nonsyndromic hearing impairment. N Engl J Med. 2002;346:243-9.
- [Google Scholar]
- GJB2 and GJB6 gene mutations found in Indian probands with congenital hearing impairment. J Genet. 2009;88:267-72.
- [Google Scholar]
- Absence of GJB6 mutations in Indian patients with non-syndromic hearing loss. Int J Pediatr Otorhinolaryngol. 2011;75:356-9.
- [Google Scholar]





