Translate this page into:
Molecular characterization of human Dirofilaria isolates from Kerala
Reprint requests: Dr Bindu Lakshmanan, Department of Veterinary Parasitology, College of Veterinary & Animal Sciences, Mannuthy, Thrissur 680 651, Kerala, India e-mail: bindul@kvasu.ac.in
-
Received: ,
This is an open access article distributed under the terms of the Creative Commons Attribution-NonCommercial-ShareAlike 3.0 License, which allows others to remix, tweak, and build upon the work non-commercially, as long as the author is credited and the new creations are licensed under the identical terms.
This article was originally published by Medknow Publications & Media Pvt Ltd and was migrated to Scientific Scholar after the change of Publisher.
Abstract
Background & objectives:
Human dirofilariosis is a well-recognized zoonosis caused by several species of the genus Dirofilaria. The disease is prevalent among canines and human beings in Kerala. The objective of the present study was to confirm the human Dirofilaria isolates by molecular characterization.
Methods:
The worms or segments obtained from human sources were subjected to diagnostic polymerase chain reaction (PCR) targeting the cytochrome oxidase subunit 1 (COI) and 5S ribosomal RNA (rRNA) genes of Dirofilaria repens. The amplicons were sequenced and analyzed.
Results:
The filariid nematodes recovered from ocular as well as subcutaneous tissue of human origin were identified as D. repens based on PCR targeting COI as well as 5S rRNA genes. The phylogenetic analysis of the COI gene nucleotide sequence obtained in the present study showed that D. repens shared the closest evolutionary relationship with D. honkongensis.
Interpretation & conclusions:
Molecular identification of D. repens isolated from human source assumes significance from the point of zoonotic threat of this mosquito-borne nematode. Phylogenetic analysis revealed a close evolutionary relationship with Asian isolate of D. honkongensis. Timely detection and treatment of infection in dogs, together with mosquito control, should be an integral part of the control strategy of this disease.
Keywords
5S ribosomal RNA gene
cytochrome oxidase subunit 1 gene
Dirofilaria repens
dirofilariosis
human
polymerase chain reaction
Dirofilaria species infect a wide range of domestic and wild animals. Dirofilaria immitis and D. repens are the most important species associated with cardiopulmonary and subcutaneous dirofilariosis in canines, felines and other carnivores. These two species of canine origin are the most important aetiological agents attributed to cause accidental infection in human beings1. Humans can also be infected by other non-canine species including D. tenuis (from racoon), D. ursi (from bears), D. subdermata (from porcupines) and D. striata from (bobcats) occasionally1. A novel Dirofilaria species D. honkongensis has also been reported from humans and dogs in Hong Kong1.
Kerala is considered as a focus of human dirofilariosis though zoonotic dirofilariosis has been reported from Karnataka, Assam and Odisha2. D. repens is the most prevalent canine species in the State associated with subcutaneous granulomas and pruritus in dogs. Prevalence of canine D. repens infection was reported to be 7-24 per cent in Kerala3. Human infections are dependent directly on mosquito density and abundance of microfilaraemic dogs in a particular geographic area. In human beings, apart from ocular infections, subcutaneous and pulmonary nodules, are the most widely reported clinical manifestations1.
Human infections are usually diagnosed based on morphological features of the worm and histological studies of the tissue sections. However, conventional histological identification is often limited to genus level and requires considerable expertise. Alteration in parasite structures and decomposition of worms inside nodules lead to ambiguity in species identification based on morphology and anatomic locations alone. Thus, the use of diagnostic techniques with reduced predictive value is inadequate to ascertain the causative Dirofilaria spp4. Genetic analysis along with morphological study is imperative to confirm the identity of the Dirofilaria spp5. The present study was undertaken to attempt molecular characterization of Dirofilaria spp. obtained from human sources in Kerala.
Material & Methods
Eight worms/segments recovered from the eye (n=1) and subcutaneous nodules (n=7) of patients were presented to the department of Veterinary Parasitology, College of Veterinary and Animal Sciences, Mannuthy, Thrissur, for identification from Jubilee Mission Medical College and Research Institute, Thrissur, during June 2013 to June 2015. A preliminary morphological identification was performed based on morphological features of the entire worms6. The identified nematodes along with fragments of nematodes were stored in 70 per cent ethanol in −20°C until further processing for molecular studies. Entire D. repens male nematodes recovered from subcutaneous nodule of dogs presented at University Veterinary Hospital, Mannuthy, were also stored to be used as known positive template. No template control as well as DNA from an unrelated trematode worm (Schistosoma spindale) served as negative controls.
DNA extraction: The nematodes of human and canine origin were processed for DNA extraction using QIAGEN DNeasy Blood and Tissue Kit (Qiagen, Germany) according to the manufacturer's protocol. The DNA content and purity of the final elutes were estimated using a nano spectrophotometer (NanoDrop 200 C, Thermo Scientific, USA). The concentration was determined at 260 nm and purity at 260:280 nm ratio. Samples which yielded a ratio between 1.7 and 1.9 were selected for analysis.
Polymerase chain reaction (PCR) protocol: The primer sets used for standardizing diagnostic PCR protocol are given in Table17. The PCR for amplification of mitochondrial DNA employed primers that amplify cytochrome oxidase subunit 1 (COI) gene. The reaction was performed in a 25 μl reaction volume containing 2.5 μl of buffer (10x) without MgCl2, 200 μM each of dNTPs, 25 pmol each of forward and reverse primers, 1.5 mM of MgCl2, 1U of Taq DNA polymerase and 5.0 μl of template DNA. The reagents and primers were procured from Sigma Aldrich, Bengaluru. A gradient thermal cycling programme (MJ Mini, Bio-Rad, USA) with initial denaturation at 94°C for two minutes followed by 32 cycles of denaturation (94°C, 30 sec), annealing (58-62°C, 30 sec), extension (72°C, 30 sec) and a final extension at 72°C for seven minute was adopted for the reactions with DR CO1 F1, R1 and DH internal transcribed spacer (ITS) F1, R1 primer sets. For amplification of 5S ribosomal RNA (rRNA) gene using D.rep F1, R1 primer sets, annealing temperature range of 60-70°C was used.

The amplicons were electrophoresed in 1.5 per cent agarose gel and sizes were resolved using DNA ladder (GeneRuler 1 Kb Plus, Fermentas, USA). The amplicons were purified using silica gel purification columns (GeneJET, Thermo Scientific) and sent for sequencing to SciGenom Labs Pvt. Ltd., Cochin. The sequences were aligned using Sequencher Version 5.0 (SciGenom Labs Pvt. Ltd.) were further subjected to sequence analysis using NCBI BLASTn (https://blast.ncbi.nlm.nih.gov).
The phylogenetic analysis was performed using the MEGA 6.08. Phylogenetic tree was constructed using maximum likelihood method from nucleotide sequences of the corresponding mitochondrial genes related to Dirofilaria spp. available in the Genbank (https://www.ncbi.nlm.nih.gov). In silico restriction enzyme (RE) analysis of the nucleotide sequences was performed using NEBcutter online tool version 2.0 (New England BioLabs, USA) to analyze the RE cutting pattern.
Results
The entire nematodes (n=5) were grossly white, thread-like with pointed extremities and measured an average of 102 μm (length) × 0.428 μm (width). The microscopic evaluation of the head end revealed a long oesophagus and an anterior vulval opening. The tail end was bluntly rounded and eggs could not be observed within the uterus. These morphologic and morphometric features were suggestive of immature female Dirofilaria spp. The fragments of recovered worms from three patients could not be identified morphologically.
PCR analysis: PCR analysis was done for further species confirmation. Gradient PCR using DR COI primers targeting COI gene yielded an amplicon of approximately 209 bp size specific for D. repens at an annealing temperature of 62°C (Fig. 1). This protocol was applied to other DNA samples extracted from worm fragments and was found to yield consistent results. Known positive DNA from D. repens of canine origin yielded amplicons of similar size. There was no amplification in negative controls. The sequence of partial COI gene was blasted using BLASTn tool to analyze their similarity with other published sequences available in online databases. The sequence was 99 per cent similar to the corresponding gene sequence of D. honkongensis and 98 per cent similar to several reported COI sequences of D. repens of human and canine origin isolated from different countries such as Japan, Slovakia, Germany, Poland and Italy. There was no significant similarity with any other species. Owing to the significant similarity of nucleotide sequence of the Kerala isolate with D. honkongensis, a novel Dirofilaria species described in humans and canines, amplification was also attempted with DH ITS primers sets reported to be specific for ITS1-5.8s region of D. honkongensis. Specific amplification was not observed in any of the samples tested. Amplification using D. repens-specific primers targeting 5S rRNA gene (D.rep F1, R1) produced duplet bands of approximately 153 bp and 247 bp at 69°C (Fig. 2) which further validated the species identification. This protocol was applied to other DNA samples extracted from worm fragments and was found to yield consistent results. There was no amplification in negative controls. Similar amplicons were observed from known positive DNA of canine origin as well. The partial COI gene sequence was submitted to Genbank and assigned with accession no. KT588609.
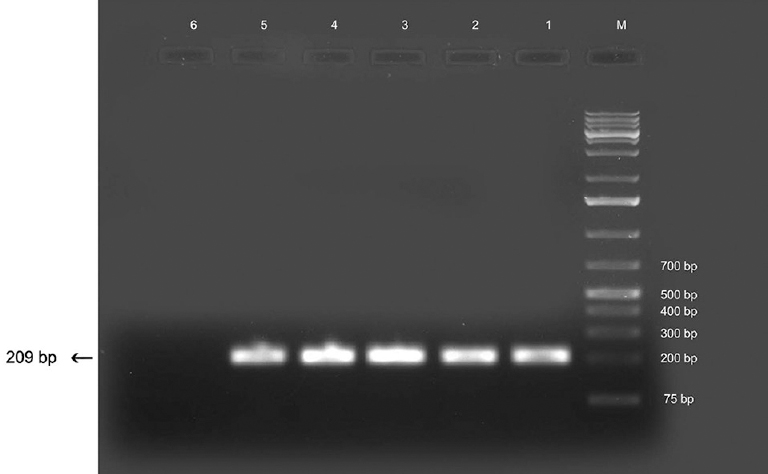
- Polymerase chain reaction products using DR COI primers. Lane M, DNA ladder; lanes 1-4, human isolates; lane 5, canine isolate; lane 6, negative control.
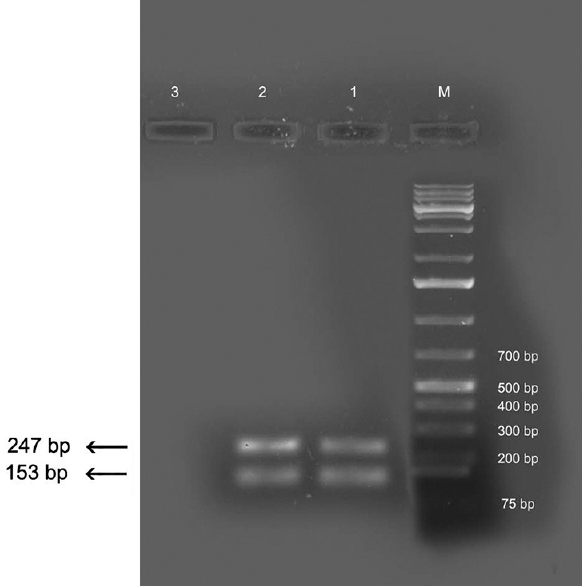
- Polymerase chain reaction products using D.rep primers. Lanes M, DNA ladder; lanes 1-2, human isolates; lane 3, negative control.
Nucleotide analysis: Sequences corresponding to COI gene of D. honkongensis (JX187591) and D. repens from different regions including Slovakia (KC985240), Poland (KM370872), Sicily (JF461458), Rome (AM749234), Germany (KF410864, KF692102), Italy (DQ358814) and Tokyo (AB 973225) available in online database were aligned using Clustal W (http://www.genome.jp/tools-bin/clustalw) with COI sequence of D. repens obtained in the present study. These aligned sequences were used to construct phylogenetic tree to analyze the evolutionary history using maximum likelihood method based on Tamura-Nei Model in Mega 6.0. The COI sequence of Ascaris lumbricoides (NC 016198) was used as the out-group for analysis. The original bootstrap tree was obtained from 1000 replicates (Fig. 3) revealed that D. repens identified in the present study shared the closest relationship with D. honkongensis. This cluster distinctly differed from the rest of the isolates including the cluster containing D. repens (Tokyo) of human origin (AB 973225).
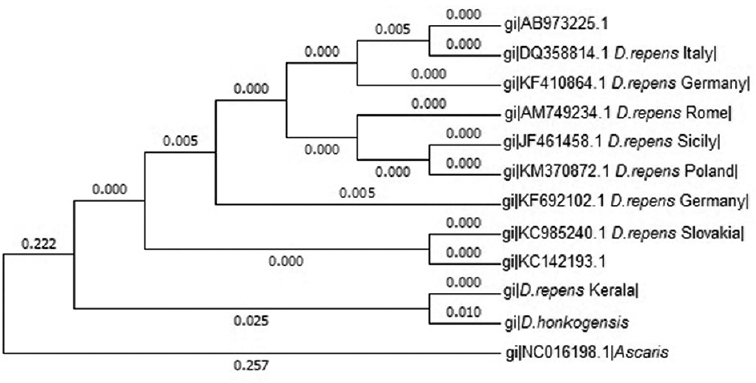
- Phylogenetic tree of Dirofilaria repens based on COI gene sequences.
The COI sequence of D. repens analyzed using the online software, NEBcutter, revealed several RE recognition sites within the sequences. The RE pattern revealed RE cleavage sites for Hpy188I, SfcI, MseI, AseI, DdeI, BspCNI, Sau3AI, BfuCI and MboI. The pattern also revealed a unique cutting site for HpyCH41V at position 114. The RE pattern was different from that of D. honkongensis. The pattern displayed more similarity to published D. repens sequence (KC 985240, AJ 271614 and KF 410864) and it could be observed that HpyCH41V and Hpy1881 cleavage sites were present in Kerala isolates only.
Discussion
Human dirofilariosis caused by D. repens910111213, D. immitis14 and D. tenuis15 has been reported from different parts of India based on morphological and morphometric features. Some cases of subcutaneous human dirofilariosis due to D. repens in Japan were mistakenly diagnosed as D. immitis infection based on morphological and serological diagnosis5. Confirmatory species identification especially of immature stages of the female Dirofilaria often demands further validation15. Mitochondrial and genomic DNA sequences were long relied upon for species confirmation of Dirofilaria in dogs, human beings as well as vector mosquitoes1617.
Mitochondrial genes are more frequently targeted for helminth identification than nuclear genes18. A PCR protocol was standardized using DR COI primers to amplify partial COI gene of Dirofilaria spp. The reaction yielded a D. repens specific product of approximately 209 bp size with the DNA from entire worms and worm fragments. These primers have been reported to effectively detect and differentiate larvae of D. immitis and D. repens in canine blood and mosquitoes in a duplex real-time PCR19. This gene was also targeted in several other studies12021 to confirm D. repens from human cases in Tokyo, Slovakia and Korea. Thus, results of partial COI gene amplification in the present study suggested that the nematodes belonged to D. repens. Moreover, the fragments of recovered worms, which could not be identified morphologically, yielded a positive PCR signal suggestive of D. repens. Positive amplification signals with DNA of adult male D. repens recovered from dogs and the absence of amplification in negative controls suggested the high specificity of the protocols standardized in the present study.
The BLASTn analysis of nucleotide sequence of partial COI gene in the present study revealed 99 per cent similarity to the COI gene sequences of D. honkongensis and 98 per cent similarity to several reported COI sequences of D. repens of human and canine origin isolated from different countries including Tokyo, Slovakia, Germany, Poland and Italy (KC985240, KM370872, JF461458, AM749234, KF410864, DQ358814, KF692102, AB 973225). The fact that the reported sequence data of D. honkongensis1 was based on amplification of COI gene with panfilarial primer sets, which would amplify the corresponding gene of all members of Filarioidea, precluded the possibility of the sequence obtained in our study to belong to D. honkongensis. Moreover, the D. repens-specific amplicons obtained with the primers targeting the 5S rRNA gene also confirmed the identity of the human isolates to be D. repens. The 5S ribosomal spacer of D. repens has been reported to possess a unique feature of yielding two amplification products with universal primers in PCR22. The duplet bands obtained with D.rep primers are specific to this species alone as per the published reports723. In addition, positive PCR signals were not obtained with specific D. honkongensis primers targeting the ITS 1-5.8S region. The positive PCR signals using mitochondrial and 5S rRNA gene targeted protocols established that all the human isolates of the present study were D. repens. This concurs with the earlier report of D. repens in Kerala based on morphologic observations3.
The phylogenetic tree revealed that D. repens, Kerala, shared the closest relationship with D. honkongensis, a novel Asian species reported from Honk Kong. No information on COI sequences of human isolates of D. repens from India is available, and the strong clustering of Kerala isolate with another Asian isolate suggested that these isolates shared a common ancestor. This clade distinctly differed from the rest of the isolates including the clade-containing D. repens of the human origin, Tokyo (AB 973225), reported from an Asian country. However, the human case reported from Tokyo was considered as imported dirofilariosis from Italy5 and was not of Asian origin. In the phylogenetic tree obtained in the present study, the sequence of Tokyo isolate shared the same clade with D. repens, Italy.
Molecular studies generate nucleotide sequence data which would not only serve to enrich the database of zoonotic parasite fauna but also provide an insight into the epidemiology of this mosquito-borne zoonosis and help in better understanding of host-parasite relationship involving canines and human beings. Molecular epidemiology of the infection can be unravelled by utilizing human patients and demographic details. Future studies need to be conducted on mosquito species prevalent in the region to ascertain the molecular evidence of D. repens in different tissues.
Acknowledgment
Authors thank the Kerala Veterinary and Animal Sciences University, Pookode, Wayanad, for the facilities provided and to Dr Sathyavati, Professor and Head, Department of Microbiology, Jubilee Mission Medical College and Research Institute, Thrissur, for providing the parasite specimens.
Conflicts of Interest: None.
References
- A novel Dirofilaria species causing human and canine infections in Hong Kong. J Clin Microbiol. 2012;50:3534-41.
- [Google Scholar]
- Human and animal dirofilariasis: The emergence of a zoonotic mosaic. Clin Microbiol Rev. 2012;25:507-44.
- [Google Scholar]
- Molecular analysis of Dirofilaria repens removed from a subcutaneous nodule in a Japanese woman after a tour to Europe. Parasite. 2015;22:2.
- [Google Scholar]
- Helminths, arthropods and protozoa of domesticated animals. London: Bailliere Tindall; 1982.
- Discrimination between six species of canine microfilariae by a single polymerase chain reaction. Vet Parasitol. 2006;135:303-14.
- [Google Scholar]
- MEGA6: Molecular Evolutionary Genetics Analysis Version 6.0. Mol Biol Evol. 2013;30:2725-9.
- [Google Scholar]
- Subcutaneous dirofilariasis in Southern India: A case report. Ann Trop Med Parasitol. 2005;99:437-40.
- [Google Scholar]
- Subcutaneous human dirofilariasis due to Dirofilaria repens: Report of two cases. J Glob Infect Dis. 2011;3:199-201.
- [Google Scholar]
- Human pulmonary dirofilariasis in India: A case report. J Trop Med Hyg. 1989;92:425-6.
- [Google Scholar]
- Molecular survey of Dirofilaria immitis and Dirofilaria repens by direct PCR for wild caught mosquitoes in the republic of Korea. Vet Parasitol. 2007;148:149-55.
- [Google Scholar]
- Highly sensitive multiplex PCR for simultaneous detection and discrimination of Dirofilaria immitis and Dirofilaria repens in canine peripheral blood. Vet Parasitol. 2010;172:160-3.
- [Google Scholar]
- Dirofilaria repens in Vietnam: Detection of 10 eye and subcutaneous tissue infection cases identified by morphology and molecular methods. Korean J Parasitol. 2012;50:137-41.
- [Google Scholar]
- A duplex real-time polymerase chain reaction assay for the detection of and differentiation between Dirofilaria immitis and Dirofilaria repens in dogs and mosquitoes. Vet Parasitol. 2012;185:181-5.
- [Google Scholar]
- Histological and molecular confirmation of the fourth human case caused by Dirofilaria repens in a new endemic region of Slovakia. J Helminthol. 2013;87:85-90.
- [Google Scholar]
- A rare human case of Dirofilaria repens infection in the subcutaneous posterior thorax with molecular identification. Korean J Parasitol. 2015;53:329-33.
- [Google Scholar]
- 5S ribosomal spacer sequences of some filarial parasites: Comparative analysis and diagnostic applications. Mol Cell Probes. 2000;14:285-90.
- [Google Scholar]
- Unusual organization of the 5S ribosomal spacer in Dirofilaria repens: Absence of a canonical spliced leader 1 sequence. Parasitol Res. 2000;86:497-9.
- [Google Scholar]






