Translate this page into:
Identification of a single nucleotide polymorphism indicative of high risk in acute myocardial infarction
Reprint requests: Dr Kavita Shalia, Sir H. N. Medical Research Society, Court House, L. T. Road, Mumbai 400 002, Maharashtra, India e-mail: kavita.shalia@rfhospital.org
-
Received: ,
This is an open access article distributed under the terms of the Creative Commons Attribution-NonCommercial-ShareAlike 3.0 License, which allows others to remix, tweak, and build upon the work non-commercially, as long as the author is credited and the new creations are licensed under the identical terms.
This article was originally published by Medknow Publications & Media Pvt Ltd and was migrated to Scientific Scholar after the change of Publisher.
Abstract
Background & objectives:
Acute myocardial infarction (AMI) is a major health concern in India. The aim of the study was to identify single nucleotide polymorphisms (SNPs) associated with AMI in patients using dedicated chip and validating the identified SNPs on custom-designed chips using high-throughput microarray analysis.
Methods:
In pilot phase, 48 AMI patients and 48 healthy controls were screened for SNPs using human CVD55K BeadChip with 48,472 SNP probes on Illumina high-throughput microarray platform. The identified SNPs were validated by genotyping additional 160 patients and 179 controls using custom-made Illumina VeraCode GoldenGate Genotyping Assay. Analysis was carried out using PLINK software.
Results:
From the pilot phase, 98 SNPs present on 94 genes were identified with increased risk of AMI (odds ratio of 1.84-8.85, P=0.04861-0.003337). Five of these SNPs demonstrated association with AMI in the validation phase (P<0.05). Among these, one SNP rs9978223 on interferon gamma receptor 2 [IFNGR2, interferon (IFN)-gamma transducer 1] gene showed a significant association (P=0.00021) with AMI below Bonferroni corrected P value (P=0.00061). IFNGR2 is the second subunit of the receptor for IFN-gamma, an important cytokine in inflammatory reactions.
Interpretation & conclusions:
The study identified an SNP rs9978223 on IFNGR2 gene, associated with increased risk in AMI patient from India.
Keywords
Acute coronary syndrome
cardiovascular disease
coronary artery disease
genetic variations - microarray
Coronary heart disease (CHD) is a major health problem in India1. Of particular concern is the incidence of coronary artery disease (CAD) in the young reported as 12-16 per cent in Indians, higher than several other ethnic groups23. Clinical manifestations of CAD encompass a spectrum of clinical entities, ranging from stable angina, unstable angina (UA), non-ST-segment elevation myocardial infarction (NSTEMI) to ST-elevation myocardial infarction (STEMI)2, and the latter three conditions cause acute coronary syndrome (ACS). The term acute myocardial infarction (AMI) is used when there is evidence of myocardial necrosis in a clinical setting consistent with myocardial ischaemia4.
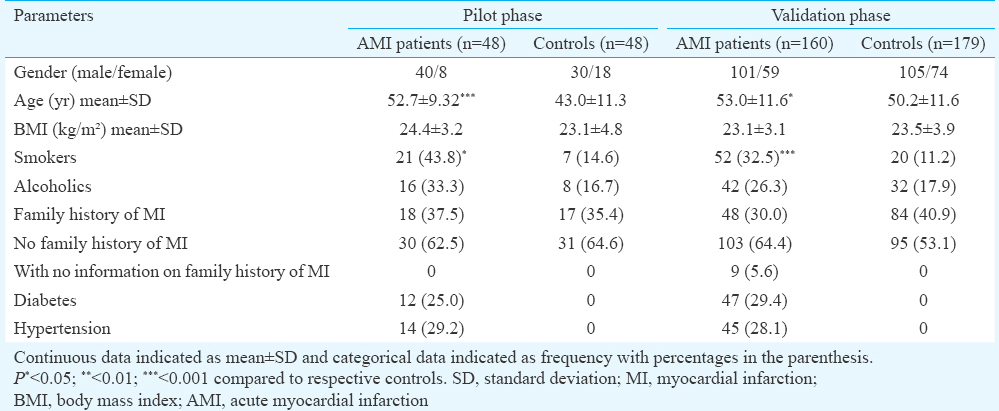
The conventional risk factors for AMI are age, smoking, hypertension, diabetes, dyslipidaemia, abdominal obesity, etc5. While lifestyle clearly plays a role in these risk factors, a large proportion of the inter-individual variability is due to inherited factors6. Thus, it is imperative to understand the genomic milieu of the individuals developing AMI which may help in identifying an individual's susceptibility to the disease.
Genetic variants as single nucleotide polymorphisms (SNPs) occur at 300-1000 bases along the entire human genome, in the coding and non-coding regions78. Thus, constructing SNP maps enables the identification of panel of SNPs and associated multiple genes as predicting biomarkers for high risk of AMI in individuals. Several reports of genome-wide association studies (GWASs) for common diseases including Alzheimer's disease, asthma, Crohn's disease, diabetes type 1 or 2, hypertension, obesity and AMI have been reported9. Various loci and genetic variants have been documented associated with the risk of myocardial infarction10111213. Among these, association of a variant locus in a non-coding region at 9p21.3 near CDKN2A (encoding the prototypic INK4 protein p16INK4a) and CDKN2B (encoding p15INK4b) genes with AMI was a common finding10111213. This locus was initially implicated in CAD14. CDKN2A and CDKN2B genes play an important role in the regulation of cell cycle and in transforming growth factor-β-induced growth inhibition which influences the pathogenesis of atherosclerosis14. Besides high-risk SNPs on chromosome 9p21, differential expression of the genes has also been documented with increasing severity of atherosclerosis1011. On the Indian scenario, however, SNPs at locus 9p21 and its flanking region have not been reported to be associated with AMI so far15.
With multiple genomic targets potentially contributing to disease, analysis requires the use of flexible, accurate tools. High-throughput genomic technologies enable a deeper understanding of disease aetiology at a molecular level. Microarray is one of such techniques, which is used to sift through and analyze the genetic variations contained within a genome.
GWAS of cardiovascular disease (CVD) suggests that genetic variants associated with CVD have low penetrance implying study in large sample sizes16. To enable this in smaller sample size, focussed SNP chips have been prepared for CVD. The Illumina HumanCVD (CVD55K) BeadChip (Illumina Inc., San Diego, USA) features 48,472 SNPs to capture genetic diversity across 2100 genes associated with CVD. BeadChip content has been derived from published scientific literature, CVD pathway analysis and recent whole-genome analysis data sets17. This rationally selected marker set offers a comprehensive coverage of genes in primary and secondary vascular disease processes including blood pressure, insulin resistance, metabolic disorders, dyslipidaemia and inflammation17. With a view to define association of specific SNPs with AMI in the current study, a Phase I (pilot phase) genetic association study was undertaken using CVD55K BeadChip and Illumina Infinium II Genotyping Assay protocol on high-throughput microarray platform, and further, in Phase II (validation phase), SNPs associated with AMI in Phase I were validated using custom-based Illumina VeraCode GoldenGate Genotyping Assay, in patients with AMI.
Material & Methods
During the study period, consecutive AMI patients (n=236) with prolonged chest pain for more than 30 min, ST-segment elevation of >0.5 mV on two adjacent ECG leads and elevated cardiac enzymes were included in the study. Exclusion criteria were valvular heart disease, known cardiomyopathy, malignancy and renal or liver diseases. Controls (n=244) comprised healthy individuals (participants in health check-up camps) with systolic/diastolic blood pressure=135/85 mmHg or less, normal carotid Doppler and no risk factors of CAD or clinical symptoms of any other organic disease. Demographic status, clinical history, family history and medications of the patient and control group were recorded. Criteria (arbitrary) for smoking were one cigarette a day and consumption of alcohol once a week. All participants provided written informed consent. The study was approved by the Institutional Ethics Committee of Sir H. N. Hospital and Research Centre, Mumbai. The entire study involving selection of participants at Sir H. N. Medical Research Society (Mumbai, Maharashtra), experimental and analysis of the data of the pilot phase (Dhirubhai Ambani Reliance Life Sciences Centre, Navi Mumbai, Maharashtra), validation phase (Sandor Proteomics Pvt. Ltd., Hyderabad, Andhra Pradesh) and analysis of the data of the validation phase (Clevergene Biocorp Pvt. Ltd. and Bionivid Technology Pvt. Ltd., Bangalore, Karnataka) was carried out in the span of five years from January 2009 to December 2014.
Genotyping: Genotyping of multiple SNPs was done using SNP array, a type of DNA microarray used to detect polymorphisms within a population. A microarray consists of different nucleic acid probes that are chemically attached to a substrate which can be a microchip, a glass slide or a microsphere-sized bead. The basic principles of SNP array are the convergence of DNA hybridization, fluorescence microscopy and solid surface DNA capture. The three mandatory components of the SNP arrays thus are an array containing immobilized allele-specific oligonucleotide probes, fragmented nucleic acid sequences of target fluorescent dyes and a detection system that records and interprets the hybridization signal. Usually, two probes are used for each SNP position to detect both alleles.
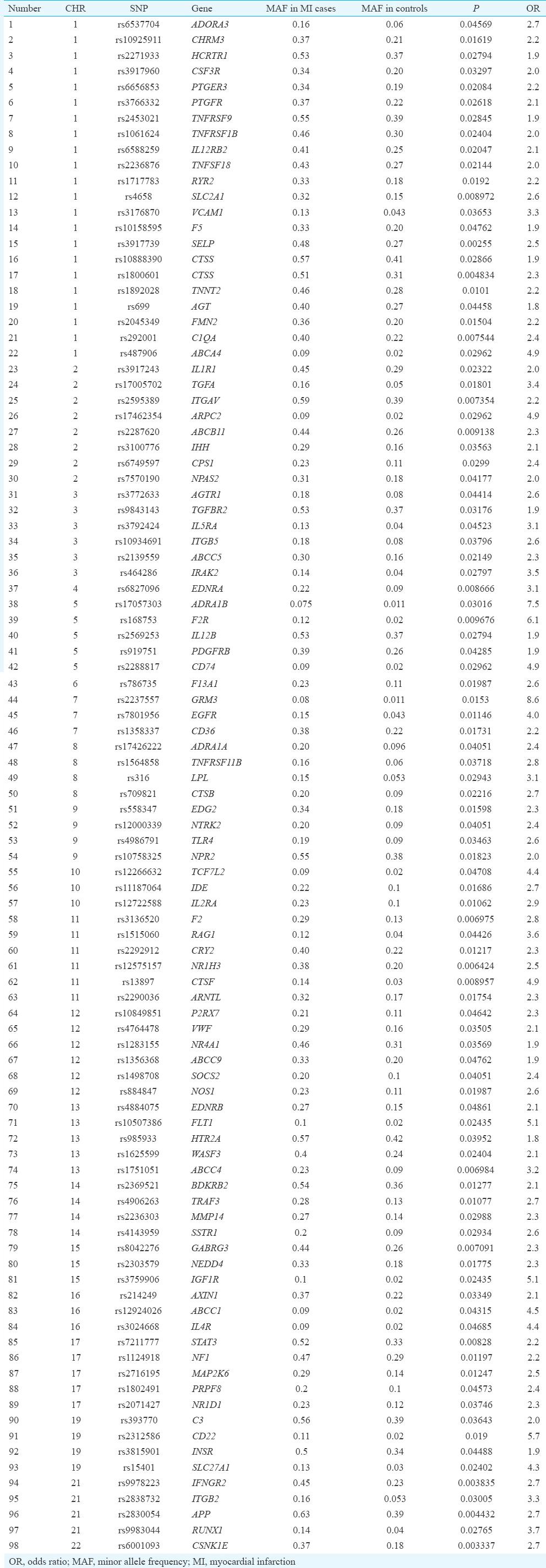
Pilot phase: In the pilot phase, 48 AMI patients and 48 controls were genotyped for AMI-associated SNPs using Illumina HumanCVD (CVD55K) BeadChip with 48,742 SNPs probes by microarray. Illumina Infinium II Genotyping Assay protocol18 was implemented as per the manufacturer's guidelines. Intensities of the beads’ fluorescence were detected using Bead Array Reader (Illumina Inc.). Genotypes for each sample were called using Illumina's inbuilt BeadStudio software version 3.3.7. The raw SNP data were exported chromosome-wise from BeadStudio software and analyzed for clustering and individual genotype call. An overall call rate of 98 per cent was provided and samples below these call rates were excluded from the analysis to avoid false-positive association. The analysis of this phase demonstrated 87 SNPs significantly associated with AMI with odds ratio (OR) of 2.0 and above. In addition, 11 SNPs with OR 1.8-<2.0 along with the identified 87 SNPs (total 98 SNPs) were validated in another independent set of AMI patients and controls.
Validation phase: Custom-based Illumina VeraCode GoldenGate Genotyping Assay19 was used to validate the SNPs associated with AMI identified at the pilot phase, in another set of AMI patients (n=188) and controls (n=196). In the validation phase, 98 identified SNPs associated with AMI from the pilot phase and 96 control SNPs were sent to Illumina for scoring and were further selected for assay based on final score, designability, rank and failure code. Of the 98 AMI-specific SNPs, two were identified with lower score of <0.4 and were excluded from the assay, and finally, 96 AMI-specific SNPs and 96 control SNPs were used to prepare the Oligo Pool Assay. The Illumina microarray platform, BeadXpress Reader (Illumina Inc.) was used for identification of microbead code and detection of fluorescent signal. The Illumina GenomeStudio V2011.1, Genotyping module version: 1.9.4 was used to obtain automated genotype clustering and calls. Samples with call rate >98 per cent were considered for analysis. Multiple sample-dependent and sample-independent quality control measures were implemented. Haploview software (https://www.broadinstitute.org/haploview) was used for estimating linkage disequilibrium (LD) between the markers and for testing for haplotypic associations. Of the 188 patients and 196 control samples analyzed, 28 patients and 17 controls were excluded for >5 per cent missing data. Thus, validation study comprised 160 patients and 179 controls, and hence, the final study population comprised 208 patients and 227 controls.
Statistical analysis: Results were expressed as frequency and percentages and mean±standard deviation for parametric variables. The analysis of significance of difference between parametric variables between two groups was performed by Student's unpaired t test. Analysis of categorical data between two groups was carried out using Chi-square test. Analyses were performed using statistical software SPSS (version 21.0, Chicago, IL, USA). PLINK software was used to carry out the statistical analysis of the microarray data, which is a whole-genome association analysis toolset indicating a binary test outcome of the presence or absence of specific alleles20.
Results
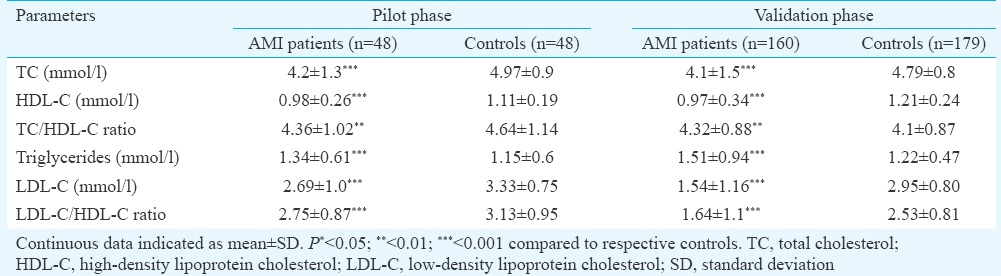
The demographic and clinicopathological data as well as lipid profile of the study population are detailed in Tables I and II, respectively. Briefly, the age range of the groups was from 35 to 65 years. In the pilot phase, there were 12 (25.0%) diabetics, 14 (29.2%) hypertensives and 21 (43.8%) smokers in the AMI group. In control group, there were seven (14.6%) smokers. The study population of validation phase comprised 160 AMI patients and 179 controls. In this study group, AMI patients with diabetes were 47 (29.4%), with hypertension 45 (28.1%) and with smoking habit 52 (32.5%). In this control group, there were 20 (11.2%) smokers.
Genotype data analysis
Pilot phase: The results of the pilot phase for individual SNPs were analyzed using PLINK. Chi-square was applied to test association of SNPs with AMI. The analysis showed 989 of the 48,742 SNPs with significantly increased minor allele frequency (MAF) in AMI cases as compared to controls (P<0.05). The advanced quality control filtering and exclusion of SNPs with OR of <1 described 869 SNPs. Illumina Genome Viewer software version 3.2.9 was used to identify location of these SNPs on chromosome and associated genes. The analysis defined 331 genes. The most biologically plausible genes associated with AMI and their roles in cardiovascular disease pathway were identified using Kyoto Encyclopedia of Genes and Genomes Pathway Database21. This analysis revealed 98 SNPs across 20 chromosomes and 94 candidate genes associated with high risk of developing AMI with OR of 1.84-8.85 and P=0.04861-0.003337 (Table III). Chromosome 1 had the maximum number of SNPs i.e. 22 SNPs; there were eight SNPs on chromosome 2, six SNPs each on chromosomes 3, 11 and 12, five SNPs on chromosome 5, 13 and 17, four SNPs on chromosomes 8, 9, 14, 19, 21, three SNPs on chromosomes 7, 10, 15, 16 and one SNP each on chromosome 4, 6 and 22.
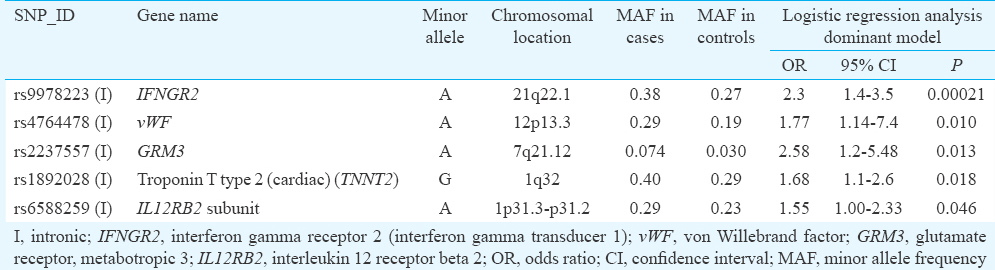
Validation phase: Ninety six of 98 identified SNPs associated with AMI in the pilot phase were further validated in another set of patients and controls. In the validation phase, of the 96 AMI SNPs, those with >5 per cent missing genotypes (six SNPs) and SNPs with MAF <0.05 (six SNPs) were excluded, and the high-quality markers were tested for violation of Hardy–Weinberg equilibrium (HWE) using goodness of fit Chi-square test. Further, two SNPs were rejected because of the distribution of their alleles violated HWE with a significant P value (P<0.001) in the control population. Thus, the quality tests resulted in a total of 82 SNPs for further association analysis. PLINK software was used to perform logistic regression to test association of SNPs with AMI by taking phenotype i.e. disease as a dependent variable and SNP as an independent variable. Allelic and genotypic associations with dominant and recessive models were applied to select the best model for performing the analysis. Thus, from 82 AMI-specific SNPs identified in the pilot phase, in dominant genetic model, five SNPs (rs9978223, rs4764478, rs2237557, rs1892028 and rs6588259) spanning four chromosomes and five genes demonstrated consistent reproducible association with AMI. The OR for five SNPs ranged from 1.55 to 2.58 and P values ranged from 0.0464 to 0.00021 (Table IV). Bonferroni corrected P value was used as cut-off for correcting of multiple testing. Significant P value threshold (Bonferroni corrected P value) for the study considering 82 unique markers was estimated as 0.00061. Therefore, of these five SNPs, only one SNP rs9978223 on interferon gamma receptor 2 [IFNGR2, interferon (IFN)-gamma transducer 1] gene showed a significant association with AMI in a dominant genetic model (OR=2.3, 95% confidence interval 1.48-3.57, P=0.00021). No SNP pairs were found to be in tight linkage disequilibrium with each other during haplotypic analysis (using Haploview software).
Discussion
AMI, a multifactorial-polygenic disease, remains a major cause of morbidity and mortality in India. The panel of genomic variants in an individual may contribute a critical risk factor identifying individuals with increased susceptibility to the disease. In this study 48 AMI patients and 48 healthy controls were screened for 48,742 human CVD SNPs. The Human CVD BeadChip has an average density of 35.5 SNPs per locus providing more than twice the density of standard whole-genome genotyping arrays17. This higher density offers greater resolution for detecting causal variants, including variants with low ORs, on a limited sample size17. This pilot phase identified 98 SNPs across 94 genes with significantly increased MAF and increased OR in the AMI patients. Of these, 96 SNPs were analyzed in additional 160 cases and 179 controls. The SNP rs9978223 in IFNGR2 on chromosome 21q22.1 was associated with AMI (P=0.00021) below Bonferroni corrected P value (P=0.00061). IFNGR2 associated with signal transduction in cells is the second subunit of the receptor for IFN-gamma, an important cytokine in inflammatory reactions. Increased expression of IFNGR2 has been observed in cardiac syndrome X associated with myocardial ischaemia22 and development of in-stent restenosis associated with tissue proliferation during neointima formation23. This SNP associated with AMI in our patients has not been documented in the Catalog of Published GWASs9. Several variants associated with CVD reported in GWAS10111213 and SNPs on CVD55K BeadChip were not observed in our study.
The location of the SNPs within the gene also influences expression and biological effect. The rs9978223 is located in the intronic region and may be associated with regulatory functions. The introns enhance expression of a gene by influencing transcription initiation, elongation, termination, polyadenylation, nuclear export and thus overall mRNA stability24.
Apart from the sample size, the limitation of the study was age difference between patients and controls. In the pilot study, the mean age of the control group was lower by 25.5 per cent. The aim of the pilot study was to narrow down the large number of SNPs of CVD55K BeadChip to those significantly associated with AMI which were then validated in larger independent groups of patient and control. In the validation phase, the mean age of the controls was lower by 5.6 per cent than patient group. Another difference was more number of smokers in the patient group as compared to control group in both the phases.
The covariate analysis was omitted. In the present case-control type of study, we had not selected a random sample of study subjects, but rather ascertain cases and controls from the source population with strict criteria. As indicated by Mefford and Witte25, this ascertainment process can create a correlation between the genetic variant and covariate in cases as they will be enriched for both risk genotypes and high-risk covariate levels. In the presence of this induced correlation, omitting the covariate from a logistic regression model may be the most powerful approach. Including the covariate could substantially increase the standard error of the genetic variant association (i.e. due to the induced correlation), resulting in a larger power loss than might arise from omitting the covariate and biasing the association towards the null hypothesis.
Genomic constitution of an individual is associated with an increase or decrease in the susceptibility to disease progression, response to treatment and recurrence of the disease26. Validation in a relatively larger group may nullify some of the data from the pilot phase executed in a smaller sample.
In conclusion, the present study identified a single SNP rs9978223 on IFNGR2 gene with substantial significance to be associated with high risk of AMI.
Acknowledgment
Authors acknowledge Dhirubhai Ambani Reliance Life Sciences Centre (Navi Mumbai) for the use of CVD55K BeadChip, the Microarray System of Illumina and funding the entire pilot phase of the project and Sir H. N. Medical Research Society for providing finance for the rest of the project. Authors also acknowledge Sir H. N. Hospital and Research Centre and Rajawadi Hospital for permitting the recruitment of the patients, Ms Rachna Mehta, Ms Poonam Pawar, Ms Siddhi Divekar, Ms Charuta Godbole for their assistance in recruiting the study participants and carrying out DNA extraction and its qualitative and quantitative analysis, and Sandor Proteomics Pvt. Ltd. for technical support in the customized GoldenGate Genotyping Assay. Authors thank Dr Tony Jose from Clevergene Biocorp Pvt. Ltd. & Bionivid Team for providing software and analytical support of the microarray data.
Conflicts of Interest: None.
References
- Acute myocardial infarction in North Kerala - A 20 year hospital based study. Indian Heart J. 1991;43:93-6.
- [Google Scholar]
- Coronary anatomy and prognosis of young, asymptomatic survivors of myocardial infarction. Am J Med. 1994;96:354-8.
- [Google Scholar]
- Joint ESC/ACCF/AHA/WHF Task Force for the Redefinition of Myocardial Infarction. Universal definition of myocardial infarction. J Am Coll Cardiol. 2007;50:2173-95.
- [Google Scholar]
- Effect of potentially modifiable risk factors associated with myocardial infarction in 52 countries (the INTERHEART study): Case-control study. Lancet. 2004;364:937-52.
- [Google Scholar]
- for International HapMap Consortium. A second generation human haplotype map of over 3.1 million SNPs. Nature. 2007;449:851-61.
- [Google Scholar]
- The NHGRI GWAS catalog, a curated resource of SNP-trait associations. Nucleic Acids Res. 2014;42:D1001-6.
- [Google Scholar]
- A common variant on chromosome 9p21 affects the risk of myocardial infarction. Science. 2007;316:1491-3.
- [Google Scholar]
- for Myocardial Infarction Genetics Consortium. Genome-wide association of early-onset myocardial infarction with single nucleotide polymorphisms and copy number variants. Nat Genet. 2009;41:334-41.
- [Google Scholar]
- Genome-wide association study identifies a susceptibility locus at 21q21 for ventricular fibrillation in acute myocardial infarction. Nat Genet. 2010;42:688-91.
- [Google Scholar]
- Genome-wide association study for coronary artery calcification with follow-up in myocardial infarction. Circulation. 2011;124:2855-64.
- [Google Scholar]
- Genomewide association analysis of coronary artery disease. N Engl J Med. 2007;357:443-53.
- [Google Scholar]
- Variant on 9p21 is strongly associated with coronary artery disease but lacks association with myocardial infarction and disease severity in a population in Western India. Arch Med Res. 2011;42:469-74.
- [Google Scholar]
- HumanCVD Genotyping BeadChip. Available from: http://www.illumina.com/documents/products/datasheets/datasheet_humancvd.pdf
- Infinium® Assay Workflow. Available from: https://www.illumina.com/Documents/products/workflows/workflow_infinium_ii.pdf
- GoldenGate ® Genotyping with VeraCode ® Technology. Available from: https://www.illumina.com/documents/products/technotes/technote_veracode_goldengate_genotyping.pdf
- [Google Scholar]
- PLINK: A tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559-75.
- [Google Scholar]
- Transcriptional activity of genes encoding interferon gamma (IFNgamma) and its receptor assessed in peripheral blood mononuclear cells in patients with cardiac syndrome X. Inflammation. 2007;30:125-9.
- [Google Scholar]
- Interferon-gamma and interferon-gamma receptor 1 and 2 gene polymorphisms and restenosis following coronary stenting. Atherosclerosis. 2005;182:145-51.
- [Google Scholar]
- The path from genome-based research to population health: Development of an international public health genomics network. Genet Med. 2006;8:451-8.
- [Google Scholar]






