Translate this page into:
New Delhi metallo-β-lactamase - type carbapenemases producing Escherichia coli isolates from hospitalized patients: A pilot study
Reprint requests: Dr. Shashi Khare, Division of Microbiology, National Centre for Diseases Control, 22-Sham Nath Marg, New 110 054, India e-mail: shashi.khare@hotmail.com
-
Received: ,
This is an open access article distributed under the terms of the Creative Commons Attribution-NonCommercial-ShareAlike 3.0 License, which allows others to remix, tweak, and build upon the work non-commercially, as long as the author is credited and the new creations are licensed under the identical terms.
This article was originally published by Medknow Publications & Media Pvt Ltd and was migrated to Scientific Scholar after the change of Publisher.
Abstract
Background & objectives:
Resistances to carbapenem group of antimicrobials among Escherichia coli due to production of carbapenemases, especially the New Delhi metallo-β-lactamase (NDM) types, pose serious challenges in the treatment of infections in healthcare settings. This study was undertaken to detect NDM producing E. coli isolates from hospitalized patients with urinary tract infection (UTI).
Methods:
A total of 30 non-repetitive isolates of E. coli from hospitalized patients with clinical suspicion of UTI were subjected to antimicrobial susceptibility testing. Screening for the production of extended-spectrum β-lactamases (ESBL) was carried out by minimum inhibitory concentration (MIC) test strip ESBL followed by phenotypic confirmation by double-disc synergy test. Phenotypic confirmation of carbapenemase production was carried out by MIC test strip metallo-β-lactamases. Molecular identification of the blaNDM gene was carried out by polymerase chain reaction (PCR) and sequencing of the amplified fragment.
Results:
Seventeen of the 30 isolates were detected as ESBL producers, of which three were found to be carbapenemase producers. NDM genes were detected by PCR followed by gene sequencing in all three isolates positive for ESBL as well as carbapenemase. The amino acid sequence of the three isolates showed complete identity to the reference sequences of NDM-1, NDM-4 and NDM-8, respectively.
Interpretation & conclusions:
Our study showed the circulation of NDM variants among the clinical isolates of E. coli that were producers of ESBL as well as carbapenemase.
Keywords
Extended-spectrum beta-lactamases
metallo-β-lactamase
New Delhi metallo-β-lactamase type 1
New Delhi metallo-β-lactamase variants
Multiple drug resistance among Escherichia coli, a common pathogen associated with urinary tract infection (UTI) in hospitalized patients1, is regarded as a major problem encountered by clinicians. This is mainly contributed by extended-spectrum β-lactamases (ESBLs) type of resistance among E. coli resulting in resistance to a myriad of antibiotics including third-generation cephalosporin. Carbapenems are powerful group of antimicrobials that are not inactivated by ESBLs and, therefore, are regarded as the treatment of choice for infections by ESBL producers2. However, recent reports of resistance to carbapenem due to carbapenemase production have posed serious challenges in the treatment of such infections3. The New Delhi metallo-β-lactamase (NDM) and closely related enzymes, which are zinc-requiring metallo-β-lactamases (MBLs), capable of hydrolyzing all penicillins, cephalosporins and carbapenem group of antimicrobials, are among the most recently identified carbapenemases. The gene that encodes NDM is called blaNDM gene and has been identified on bacterial chromosomes and plasmids3. The present study was carried out to detect and analyze NDM producing isolates of E. coli obtained from hospitalized patients suffering from UTI in a tertiary care hospital in north India.
Material & Methods
The study included a total of 30 non-repetitive isolates of E. coli from 66 urine samples randomly selected from the daily urine samples referred from clinically suspected cases of UTI admitted in the Intensive Care Unit of Ram Manohar Lohia Hospital, a tertiary care hospital in New Delhi, India, between May and July 2012. Identification of E. coli isolates was based on culture and biochemical characteristics4.
The samples were transported to the Division of Microbiology, National Centre for Diseases Control (NCDC), New Delhi. The study was approved by the NCDC ethics committee.
Antimicrobial susceptibility testing and determination of minimum inhibitory concentration (MIC) for carbapenems: Antimicrobial susceptibility testing was carried out by the Kirby-Bauer method5 and the results were interpreted as per the Clinical Laboratory Standards Institute (CLSI) guidelines6. The antimicrobial discs were commercially procured (Becton Dickinson, USA). The antimicrobial discs used were meropenem (MEM-10 μg) and imipenem (IPM-10 μg). In addition, ampicillin (10 μg), co-trimoxazole (25 μg), amikacin (30 μg), gentamicin (10 μg), nitrofurantoin (300 μg), norfloxacin (10 μg), cefoxitin (30 μg), cefotaxime (30 μg), ceftazidime (30 μg), cefepime (30 μg) and amoxicillin-clavulanic acid (20/10 μg) discs were also employed. The zone of inhibition was measured and interpreted as per the CLSI guidelines.
Minimum inhibitory concentration (MIC) for carbapenems was determined by commercial MIC test strip containing gradient of antimicrobial concentrations of meropenem and imipenem from 0.002 to 32μg/ml (Liofilchem, Italy; www.liofilchem.net). MIC was determined based on CLSI breakpoint for meropenem and imipenem considering MIC (μg/ml) ≥1 as susceptible, >1 to <4 as intermediate and ≥4 as resistant6.
Screening for ESBL producers: The screening of all the E. coli isolates for ESBL production was carried on the basis of resistance to cephalosporin by Kirby-Bauer method5. These isolates were further screened by MIC test strip ESBL (Liofilchem, Italy) followed by phenotypic confirmation by double-disc synergy test (DDST)7. DDST was carried out by placing four discs, viz., cefoxitin (30 μg), cefotaxime (30 μg), ceftazidime (30 μg) and cefepime (30 μg), radially at a distance of 20 mm each from a disc containing amoxicillin-clavulanic acid (20/10 μg) on a lawn culture of the E. coli isolate on Mueller-Hinton Agar (MHA) plate. The plates were incubated aerobically at 37°C. The isolates were confirmed as ESBL producer, if the zone size around any of the discs was enhanced towards amoxicillin-clavulanic acid disc789. K. pneumoniae ATCC 700603 (ESBL producer) strain was used as positive control.
Screening for metallo-β-lactamase (MBL) type carbapenemase production: This was carried out using commercial MIC test strip MBL (Liofilchem), one of the methods commonly employed for presumptive screening of MBL producing strains10. The range of antimicrobials in MBL strip had imipenem (IMI) gradient at one end (4-256 μg/ml) and gradient of imipenem (1-64 μg/ml) plus a constant level of EDTA (4 μg/ml) at other end (IMD). The isolate was considered as MBL producer if MIC ratio of (IMI/IMD) was ≥810.
Detection of New Delhi metallo-β-lactamase (NDM) gene: The isolates showing evidence of NDM production in MIC test strip MBL were further subjected to detection of blaNDM gene by PCR using the pre published sequences, forward 5’-ACCGCCTGGACCGATGACCA-3’ and reverse 5’-GCCAAAGTTGGGCGCGGTTG -3’ which amplified 264 bp fragment of the blaNDM gene11. PCR products of all the positive isolates were subjected to sequencing. The amplicons from the positive isolates were purified by PCR purification kit (QIAGEN, Hidden, Germany) and sequenced on ABI PRISM 3130XL sequencer (Applied Biosystems, USA) using Big Dye Terminator cycle sequencing kit (Perkin Elmer). In the sequences so obtained, the accuracy of the base calling with the chromatogram peaks was checked using BIOEDIT software (http://www.mbio.ncsu.edu/bioedit/bioedit.html) and edited wherever necessary, followed by blast at the National Center for Biotechnology Information website (http://www.ncbi.nlm.nih.gov/blast). The derived sequences were aligned with reference sequences from the database of GenBank. The derived sequences were submitted to the GenBank and accession numbers were obtained.
Results
A total of 30 E. coli isolates from 30 UTI patients in Delhi were screened for drug resistance. The age group of these UTI patients ranged from 21 to 70 yr (mean 41±11 yr) with male:female ratio as 1:2.
Overall resistance pattern of E. coli isolates against various antimicrobials were as follows: ampicillin (100%), co-trimoxazole (73.3%), gentamicin (53.3%), amikacin (46.7%), nitrofurantoin (43.3%), norfloxacin (33.3%) amoxicillin-clavulanic acid (43.3%), cefoxitin (56.7%), cefotaxime (46.7%), ceftazidime (56.7%), cefepime (56.7%), imipenem (10%) and meropenem (10%).
Resistance pattern for carbapenems: Analysis of isolates of E. coli by disc diffusion and MIC test strip determination revealed three of the 30 isolates as resistant to both meropenem and imipenem and additionally three as intermediate to both the antimicrobials.
Screening for ESBL production by MIC test strip ESBL: Of the 30 isolates of E. coli, 17 were phenotypically ESBL producers by MIC test strip method while three were found to be resistant to meropenem and imipenem. However, three more isolates were in the category of intermediate resistant to these two antimicrobials.
Phenotypic confirmation of ESBL production: All the 17 (56.7%) E. coli isolates screened positive for ESBL by disc diffusion and MIC test strip were confirmed to be ESBL producers by DDST. These 17 isolates were resistant to cefepime, thus ruling out AmpC production. AmpC production in the remaining 13 (43.3%) isolates could not be ruled out.
Screening for production of MBL type carbapenemase: Of the six isolates that were either intermediately resistant or resistant to carbapenems (both meropenem and imipenem) by disc diffusion and MIC test strip, only three were further identified as MBL producers by MIC test strip MBL and the other three intermediate resistant isolates did not show any MBL production (Table).
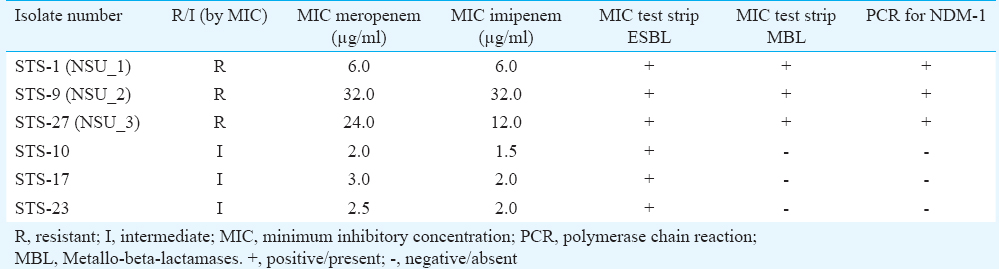
Identification of blaNDM gene: Of the six isolates that were intermediate or resistant to both meropenem and imipenem on the basis of disc diffusion and MIC test strip as well as by MIC test strip MBL, blaNDM gene was detected in the three resistant isolates only that were confirmed as NDM producers by PCR method (Table). This reconfirmed that intermediate isolates were not MBL producers.
Molecular categorization of blaNDM gene: The partial nucleotide sequence of first isolate (NSU_1, Accession number JX292120.1) showed complete identity with the sequence of reference strain NDM-1. Among the remaining two isolates, one (NSU_2, Accession number JX292121.1) showed an amino acid substitution at 154 (Met →Leu) with 100 per cent identity to the NDM-4 reference sequence and the other (NSU_3, Accession number KJ410407) showed mutations at amino acid 130 (Asp→Gly) and at 154 (Met→Leu), identical to the reference sequence of NDM-8 (Figure).

- Molecular categorization of blaNDM gene: The Plot dot identities of the derived sequence from the study (NSU-1, 2 and 3) and published New Delhi metallo-ß-lactamase-4 (NDM-4) (Accession number: WP032492624) sequence, NDM-8 (Accession number: JQ 348841) sequence from the database against NDM-1 (Accession number: AHY37787).
Discussion
In the present study, ESBL production was seen in 17 of the 30 E. coli isolates. Multiple surveys have shown the rate of ESBL production among E. coli to be highest in India (80%), followed by China (60%), while rates are lower elsewhere in East and Southeast Asia (<30%) and other countries such as Europe, Australia and North America (range 5-10%)121314. There has been a steady rise in the prevalence of ESBL producing E. coli in India, the reported prevalence increased from 18 to 40 per cent during 2003 to 2008 while it rose to 40-75 per cent during 2009 to 20122121314.
Reports prior to 2006 indicated that most E. coli isolates were sensitive to carbapenems. Studies carried out by Akram et al15 (2002 to 2006) and Padmini and Appalaraju16 (2002 to 2003) in northern India reported 100 per cent susceptibility to imipenem for urinary isolates of E. coli, while Menon et al17 in their study from southern India in 2003 reported similar pattern of susceptibility for imipenem. However, subsequent reports indicated emergence of carbapenem resistance among E. coli18. In the present study, three of 30 (10%) E. coli isolates were resistant to both meropenem and imipenem. This was in accordance to studies from elsewhere in India viz., Delhi, Guwahati and Mumbai reporting resistance from 5.1 to 14 per cent for both these antimicrobials192021.
In another study from southern India, of the 4976 samples tested, 74 (1.48%) yielded multidrug resistant isolates that included 10 E. coli isolates resistant to both meropenem and imipenem22. In a study from Kashmir, of the 1625 Gram-negative isolates, 6.0 per cent were resistant to both meropenem and imipenem23. In a hospital based study on neonatal septicemia cases from Kolkata, India, 105 (37%) of the 285 samples yielded isolates identified as Enterobacteriaceae, including 27 E. coli isolates with the resistance rate for imipenem as 0 per cent in 2007, 11 per cent in 2008, 50 per cent in 2009, 25 per cent in 2010 and 37.5 per cent in 201124.
Several studies from India reported high incidence of NDM-like enzyme production among the carbapenem-resistant E. coli isolates from hospitals. It has been shown that NDM producing Enterobacteriaceae, including E. coli, are widespread in India25. The patients presented with a variety of hospital and community-associated infections, with UTI being the most common clinical symptom26. Deshpande et al27 reported NDM-1 in nine E. coli isolates among 24 carbapenem resistant Enterobacteriaceae in a tertiary care centre25. Of the 74 E. coli isolates showing resistance to carbapenems, 34 were positive for blaNDM gene by PCR22. In a study from North-East India on the incidence of blaNDM gene among the clinical isolates of E. coli, of the 270 E. coli isolates from various clinical samples, during 2009-2010, NDM-1 could be detected in 14 isolates (5.2%)20.
Up till now 12 published new variants of blaNDM gene differing from each other by one or two residues have been reported from various countries2829. However, NDM-1, 4 and 8 producing isolates were found to be circulating in Delhi as shown in our study. The sequence from one isolate (NSU_1) matched with that of NDM-1. The NDM-4 is known to differ from NDM-1 by a single amino acid substitution at position 154 (Met→Leu) and the isolate (NSU_2) showed 100 per cent nucleotide identity with NDM-4 variant. This amino acid substitution is responsible for an increased hydrolytic activity of NDM-4 compared to NDM-1 towards meropenem and imipenem30. The amino acid sequence of isolate NSU_3 showed substitution at position 130 (Asp→Gly) and at 154 (Met→Leu) compared with NDM-1 which is identical to NDM-8 in accordance to the published reports28293031.
In a study by Rahman et al31, 12.3 per cent (n=13) of isolates belonging to Enterobacteriaceae family at a tertiary care hospital in northern India showed resistance or reduced susceptibility to carbapenem (imipenem or meropenem). These isolates were all positive for NDM with 13 isolates as variants of NDM-1 i.e. NDM-5(2), NDM-6(8) NDM-7(3)31.
In conclusion, the present study highlighted the continued threat by ESBL producing E. coli in the hospital setting. The detection of NDM variants, i.e., NDM-1, NDM-4 and NDM-8 by the E. coli isolates warrants further exploration on a large scale in the country to estimate the prevalence of NDM producing strains in the clinical setting and to review future treatment strategies for UTIs in hospitalized patients particularly in the Intensive Care Units. The present study also indicates the importance of regular monitoring of drug resistance in the hospital for an urgent action to be taken for antibiotics stewardship in the country.
Acknowledgment
The second author (AD) received STS grant (Reference ID: 2012-01377), from the Indian Council of Medical Research, New Delhi, India.
Conflicts of Interest: None.
References
- Complicated urinary tract infection in adults. Can J Infect Dis Med Microbiol. 2005;16:349-60.
- [Google Scholar]
- Current epidemiology and growing resistance of gram-negative pathogens. Korean J Intern Med. 2012;27:128-42.
- [Google Scholar]
- Emergence of a new antibiotic resistance mechanism in India, Pakistan, and the UK: A molecular, biological, and epidemiological study. Lancet Infect Dis. 2010;10:597-602.
- [Google Scholar]
- Enterobacteriaceae: Escherichia, Klebsiella, Proteus and other genera. In: Collee JG, Fraser AG, Marmion BP, Simmons A, eds. Mackie and McCartney practical medical microbiology (14th ed). Edinburgh: Churchill Livingstone; 1999. p. :361-7.
- [Google Scholar]
- Antibiotic susceptibility testing by a standardized single disk method. Am J Clin Pathol. 1966;45:493-6.
- [Google Scholar]
- Clinical and Laboratory Standards Institute. Performance Standards for Antimicrobial Susceptibility Testing; Twenty-Third Informational Supplement. M100-S23. Wayne, PA: CLSI; 2013.
- [Google Scholar]
- Phenotypic and genotypic detection of ESBL mediated cephalosporin resistance in Klebsiella pneumoniae: Emergence of high resistance against cefepime, the fourth generation cephalosporin. J Infect. 2006;53:279-88.
- [Google Scholar]
- Extended-spectrum-beta-lactamase, AmpC, and Carbapenemase issues. J Clin Microbiol. 2010;48:1019-25.
- [Google Scholar]
- Development of TaqMan real-time polymerase chain reaction for the detection of the newly emerging form of carbapenem resistance gene in clinical isolates of Escherichia coli, Klebsiella pneumoniae, and Acinetobacter baumannii. Indian J Med Microbiol. 2011;29:249-53.
- [Google Scholar]
- Evaluation of extended spectrum beta lactamase in urinary isolates. Indian J Med Res. 2004;120:553-6.
- [Google Scholar]
- Occurrence of ESBL & Amp-C beta-lactamases & susceptibility to newer antimicrobial agents in complicated UTI. Indian J Med Res. 2008;127:85-8.
- [Google Scholar]
- Emergence of high levels of extended-spectrum-beta-lactamase-producing gram-negative bacilli in the Asia-Pacific region: Data from the Study for Monitoring Antimicrobial Resistance Trends (SMART) program, 2007. Antimicrob Agents Chemother. 2009;53:3280-4.
- [Google Scholar]
- Etiology and antibiotic resistance patterns of community-acquired urinary tract infections in J N M C Hospital Aligarh, India. Ann Clin Microbiol Antimicrob. 2007;6:4.
- [Google Scholar]
- Extended spectrum -lactamases in urinary isolates of Escherichia coli and Klebsiella pneumoniae-prevalence and susceptibility pattern in a tertiary care hospital. Indian J Med Microbiol. 2004;22:172-4.
- [Google Scholar]
- Comparison of double disc and three dimensional methods to screen for ESBL producers in a tertiary care hospital. Indian J Med Microbiol. 2006;24:117-20.
- [Google Scholar]
- Carbapenem resistance among Escherichia coli and Klebsiella pneumoniae in a tertiary care hospital in South India. Indian J Med Microbiol. 2012;30:93-5.
- [Google Scholar]
- Ganga Ram Study Finds High Levels of Superbug NDM1. The Times of India 2011 Oct 05. Available from: https://timesofindia.indiatimes.com/city/delhi/Ganga-Ram-studyfinds-high-levels-of-superbug-NDM1/articleshow/10239023.cms
- [Google Scholar]
- Incidence of bla NDM-1 gene in Escherichia coli isolates at a tertiary care referral hospital in Northeast India. Indian J Med Microbiol. 2013;31:250-6.
- [Google Scholar]
- Multidrug-resistant Enterobacteriaceae including metallo-β-lactamase producers are predominant pathogens of healthcare-associated infections in an Indian teaching hospital. Indian J Med Microbiol. 2011;29:22-7.
- [Google Scholar]
- Phenotypic identification & molecular detection of bla (ndm-1) gene in multidrug resistant Gram-negative bacilli in a tertiary care centre. Indian J Med Res. 2014;139:625-31.
- [Google Scholar]
- NDM-1 (New Delhi metallo beta lactamase-1) producing Gram-negative bacilli: Emergence & clinical implications. Indian J Med Res. 2014;140:672-8.
- [Google Scholar]
- A five-year experience of carbapenem resistance in Enterobacteriaceae causing neonatal septicaemia: Predominance of NDM-1. PLoS One. 2014;9:e112101.
- [Google Scholar]
- Increasing prevalence and dissemination of NDM-1 metallo-β-lactamase in India: Data from the SMART study (2009) J Antimicrob Chemother. 2011;66:1992-7.
- [Google Scholar]
- Emergence of New Delhi metallo-β-lactamase 1 (NDM-1) producing and multidrug resistant uropathogens causing urinary tract infections in Andaman Islands, India. Microb Drug Resist. 2013;19:457-62.
- [Google Scholar]
- New Delhi Metallo-beta lactamase (NDM-1) in Enterobacteriaceae: Treatment options with carbapenems compromised. J Assoc Physicians India. 2010;58:147-9.
- [Google Scholar]
- Worldwide dissemination of the NDM-type carbapenemases in Gram-negative bacteria. Biomed Res Int. 2014;2014:249856.
- [Google Scholar]
- NDM-8 metallo-β-lactamase in a multidrug-resistant Escherichia coli strain isolated in Nepal. Antimicrob Agents Chemother. 2013;57:2394-6.
- [Google Scholar]
- NDM-4 metallo-β-lactamase with increased carbapenemase activity from Escherichia coli. Antimicrob Agents Chemother. 2012;56:2184-6.
- [Google Scholar]
- Prevalence and molecular characterisation of New Delhi metallo-β-lactamases NDM-1, NDM-5, NDM-6 and NDM-7 in multidrug-resistant Enterobacteriaceae from India. Int J Antimicrob Agents. 2014;44:30-7.
- [Google Scholar]






