Translate this page into:
Genetic environment of OXA-2 beta-lactamase producing Gram-negative bacilli from a tertiary referral hospital
‡For correspondence: ab0404@gmail.com
This is an open-access article distributed under the terms of the Creative Commons Attribution-Noncommercial-Share Alike 3.0 Unported, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
This article was originally published by Medknow Publications & Media Pvt Ltd and was migrated to Scientific Scholar after the change of Publisher.
Sir,
OXA-2 type beta lactamses belong to Ambler molecular class D and functional Group 2d. These types of beta lactamases are characterized by their high hydrolytic spectrum of activity against cloxacillin and oxacillin, and are poorly inhibited by clavulanic acid. Presence of this gene wasfirst reported in Pseudomonas in France1, in Escherichia coli from Israel2, and in India it was reported in E. coli in 20073. However, there is no knowledge regarding genetic environment and gene location of this resistant determinant from this part of the world. Our study reports presence of blaOXA-2 within IncF plasmid in a tertiary referral hospital of north-east India.
This study was conducted in the department of Microbiology, Assam University, Silchar. A total number of 476 consecutive, non-duplicates, Gram-negative rods consisting of members of Enterobacteriaceae family and non-fermenting Gram-negative rods were isolated from different clinical specimens spanning a period of 12 months (March 2012 to February 2013) from different Wards/Clinics of Silchar Medical College and Hospital, Assam, India (Table). Screening and confirmation for extended spectrum beta lactamases (ESBLs) was done as per Clinical Laboratory Standards Institute (CLSI) guidelines4. Multiplex PCR was performed to characterize ESBL genes1. Reactions were run under the following conditions: initial denaturation 94°C for 5 min, 33 cycles of 94 °C for 35 sec, 51°C for one min, 72°C for one min and final extension at 72°C for seven min. PCR product was purified (Gene Jet Purification kit, Lithuania) and sequencing was done. For detection of class 1 and class 2 integron, integrase gene PCR was performed5. Two PCR reactions were carried out, one with HS287 and blaOXA-2 reverse, another with HS286 and blaOXA-2 forward16. The amplified products were further sequenced. Plasmids were purified by Gene Jet plasmid Miniprep kit (Thermo scientific, Lithuania). Transformation was carried out using Escherichia coli JM107 as recipient. Transformants were selected on cefotaxime (0.5 mg/l) containing Luria-Bertani agar (Hi-Media, Mumbai, India) plates. Conjugation experiments were carried out between clinical isolates as donors and a streptomycin resistant E. coli recipient strain B (Genei, Bangalore), transconjugants were selected on cefotaxime (0.5 mg/l) and streptomycin (800 mg/l) agar plates. For plasmid profiling, 1.5 μl of each sample was used and analyzed by agarose gel electrophoresis (1% agarose, Hi-Media, Mumbai, India), gel was run at 40V for 8 h at 18°C. PCR based replicon typing was carried out targeting 18 different replicon types, to perform five multiplex and three simplex PCRs as described previously7. Antimicrobial susceptibility was determined by Kirby Bauer disc diffusion and minimum inhibitory concentration (MIC) method4. Typing of isolates was done by enterobacterial repetitive intergenic consensus (ERIC) PCR8.
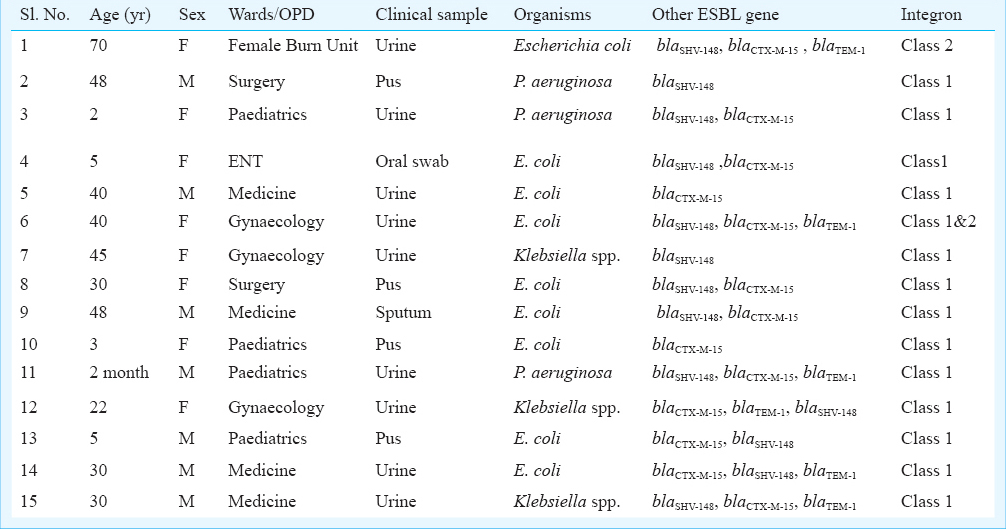
A total of 15 isolates were harbouring OXA-2 gene which was further confirmed by sequencing. Co-existence of other ESBL genes was also noticed in all 15 isolates (Table). Class 1 integron was found in 13 isolates whereas one isolate carried class 2 integron and the remaining isolate carried class 1 and 2 both (Table). Sequencing results confimed that blaOXA-2 was found to be located within class I integron in 14 isolates while presence of this gene in class2 integron could not be established. Transformation results disclosed that in 13 isolates blaOXA-2 was located within the 20 kb plasmid which was also conjugatively transferable in E. coli strain B. Incompatibility typing of plasmids demonstrated that diverse Inc group types namely I1/Iγ, FIA, FIB, FIC, Y, FrepB, K and B/o were present in all blaOXA-2 harbouring isolates. But plasmid IncF was found to be common in all isolates as well as in their transformants and transconjugants. Tigecycline (n= 13; 86.66%) was the most effective antibiotics followed by imipenem (n=12; 80%) and meropenem (n=12; 80%). High MICs was observed against different groups of cephalosporins (≥256 μg/ml; n =15) and monobactam (≥256 μg/ml; n=15). All the OXA-2 producing isolates were clonally unrelated.
This study indicates propagation of the blaOXA-2 by horizontal gene transfer additionally facilitated by integron mediated gene capture mechanism. Presence of this rare type of ESBL gene in diverse group of organisms and its carriage in integrons may restrict therapeutic options.
References
- Identification of PSE and OXA β-lactamase genes in Pseudomonas aeruginosa using PCR restriction fragment length polymorphism. J Antimicrob Chemother. 2002;50:11-8.
- [Google Scholar]
- CTX-M-2 and a new CTX-M-39 enzyme are the major extended spectrum β-lactamases in multiple Escherichia coli clones isolated in Tel Aviv, Israel. Antimicrob Agents Chemother. 2005;49:4745-50.
- [Google Scholar]
- Detection of OXA-2 group extended-spectrum beta-lactamase-producing clinical isolates of Escherichia coli from India. J Antimicrob Chemother. 2007;60:703-4.
- [Google Scholar]
- Clinical and Laboratory Standards Institute (CLSI). Performance standards for antimicrobial susceptibility testing; 21st Informational Supplement. In: M100-S21. Wayne, PA, USA: CLSI; 2011.
- [Google Scholar]
- Identification of epidemic strains of Acinetobacter baumannii by integrase gene PCR. J Clin Microbiol. 2001;39:8-13.
- [Google Scholar]
- Gene cassette PCR: Sequence-independent recovery of entire genes from environmental DNA. Appl Environ Microbiol. 2001;67:5240-6.
- [Google Scholar]
- Identification of plasmids by PCR-based replicon typing. J Microbiol Methods. 2005;63:219-28.
- [Google Scholar]
- Distribution 201 of repetitive DNA sequences in eubacteria and application to fingerprinting of bacterial genomes. Nucleic Acid Res. 1991;19:6823-31.
- [Google Scholar]





