Translate this page into:
Circulation of hepatitis A genotype IIIA virus in paediatric patients in central India
Reprint requests: Dr P.V. Barde, Scientist C, Regional Medical Research Centre for Tribals, (ICMR), Nagpur Road, Garha,Jabalpur 482 003, India e-mail: pradip_barde@hotmail.com, pradipbarde@gmail.com
-
Received: ,
This is an open-access article distributed under the terms of the Creative Commons Attribution-Noncommercial-Share Alike 3.0 Unported, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
This article was originally published by Medknow Publications & Media Pvt Ltd and was migrated to Scientific Scholar after the change of Publisher.
Abstract
Hepatitis A virus (HAV) infection, a major cause of childhood hepatitis is transmitted by orofaecal route. Children mostly suffer with subclinical infection but may have serious clinical implications leading to hospitalization and mortality. IgM ELISA and nRT PCR were conducted on the blood samples collected from HAV suspected paediatric cases referred to the viral diagnostic laboratory in the Regional Medical Research Centre for Tribals at Jabalpur, Central India. The nRT PCR products were sequenced and phylogenetic analysis was done. Of the 195 samples tested, 41 (21%) were positive for HAV antibodies, among which 38 (92%) belonged to paediatric age group and 32 per cent of these were hospitalized. nRT PCR and sequencing confirmed the presence of HAV. Phylogenic analysis revealed circulation of genotype III A in central India. Regular serological and molecular monitoring would aid in understanding epidemiology of HAV and plan intervention strategies.
Keywords
Central India
children
genotype III A
hepatitis A
Hepatitis A virus (HAV) is transmitted by orofaecal route and is a major cause of childhood hepatitis in developing countries. Every year, about 1.5 million cases of HAV are reported world over1. High incidence of HAV is directly correlated to poor sanitation and hygiene and low socio-economic conditions2. However, with improving economic conditions, the age of seroprevalence is changing from the first to the second and third decades of life resulting in increased disease severity and altered epidemiology234. HAV has positive sense single-stranded RNA which is about 7500 bases long5. It has single serotype with six genotypes. Genotypes I-III are known to infect humans6. Genotype I is prevalent in Europe and America and genotype III is endemic in Asia7. Different genotypes (IA, IB and IIIA) with a predominance of genotype IIIA are known to be circulating in southern, western and northern India3891011. However, information regarding circulating genotype is not available from central India.
We present here the finding of clinical, serological and molecular studies on the samples referred to the viral diagnostic laboratory (VDL) for HAV diagnosis in the Regional Medical Research Centre for Tribals (RMRCT) Jabalpur, Central India, from July 2012 to June 2013.
The blood samples (2 ml) were collected at tertiary care and secondary hospitals viz. Netaji Subhash Chandra Bose Medical College and Hospital and Seth Govind Das district Hospital, Jabalpur by treating physicians, from the patients suspected of acute hepatitis. The catchment areas of these hospitals were Jabalpur city and adjoining rural areas. The patients, mainly of low socio-economic background staying in the slum areas under poor hygienic conditions (such as open field defaecation, open drainage, inadequate and common water supply) are treated in these hospitals.
The patients suspected of having acute hepatitis with symptoms such as sudden onset of icterus with fever, dark urine, malaise, anorexia and nausea, etc.1 were included in the study. The written informed consent was sought from parents/guardians of the patients. The study protocol was cleared by the RMRCT's ethical committee.
ELISA and nRT-PCR: The serum was separated and screened for hepatitis A specific IgM antibody by using commercially available HAV specific IgM ELISA Kit (General Biologicals, Germany) following the manufacturer's protocol and was stored at -70°C for later use. Twenty randomly picked IgM ELISA positive samples were subjected to nested RT PCR3 with minor modifications. Briefly, the viral RNA was isolated using QIAamp viral RNA mini kit (QIAGEN, Valencia, USA) according to the manufacturer's instructions in 40 μl extraction buffer. Super Script III one step RT PCR kit with platinum Taq HiFi (Invitrogen, CA, USA) was used for the amplification according to the manufacturer's instructions. The nested PCR products (332 bp), were extracted from the gel and sequenced (n=12) directly, using Big Dye Terminator Cycle Sequencing kit (Applied Biosystems, CA, USA) on ABI 3130 XL genetic analyzer (Applied Biosystems, USA) and three representative sequences were submitted to the Gen Bank.
The information requested in the predesigned laboratory request form was analyzed using appropriate statistical tests such as z-test and odds ratio, etc. The sequences obtained (n=9) were analyzed for their homologies using Basic Local Alignment Search Tool (BLAST) (http://blast.ncbi.nlm.nih.gov/Blast.cgi), further three representative nucleotide sequences were compared38 with 16 sequences of different genotypes of HAV available in the National Center for Biotechnology Information (NCBI) database (http://www.ncbi.nlm.nih.gov/). Multiple sequence analysis was done using CLUSTAL W software12 and p distance Neighbor Joining phylogenetic tree was generated by using MEGA, version 5 applying 1000 bootstraps12 to determine genotype and to establish nearest homologous HAV virus.
The IgM ELISA conducted on the 195 samples referred to VDL revealed that 41 (21%) patients had HAV infection. The mean age of HAV cases was 6.7 ± 5.01 yr ranging from 18 months to 23 yr. The major sufferer of HAV infection were paediatric patients [OR: 10.8 (CI 3.2-36.6) P<0.0001], and the majority (66%), were in the age group of 4-10 yr. HAV infection was also detected in three adults (Table). The children in the age group of less than three years were almost 21 times at higher risk of HAV infection (OR=21.3, P<0.0001) when compared with adults. HAV cases were equally distributed in urban and rural areas. However, 82 per cent of positive cases were from the slum areas of urban Jabalpur. Seasonal trend was observed among the HAV positive cases. Of the 91 suspected cases, in monsoon and post monsoon season (July to November), 29 (31.8%) were detected positive, whereas only 12 of 104 (11.5%) were positive during pre-monsoon season (December to June) (z value = 4.156, P=0.0005).
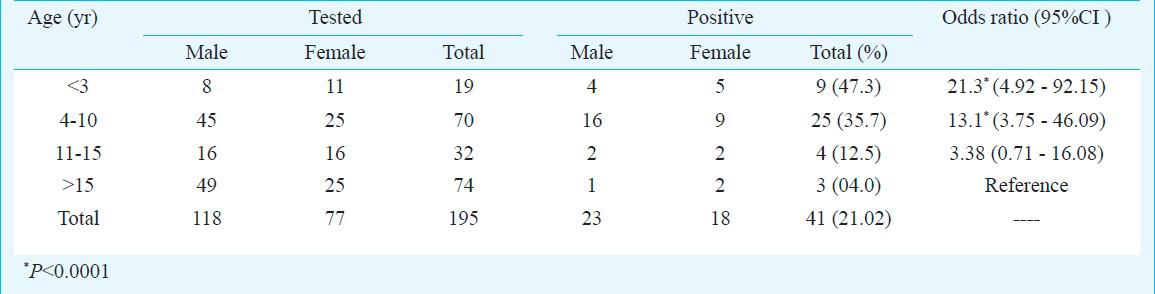
Out of 195 clinically suspected 177 cases, (91%) patients were having icterus with fever, 133 (68%) cases were suffering from anorexia, weakness with hepatomegaly, 98 (50%) were complaining of nausea and vomiting and 62 (32 %) were having liver tenderness. Thirty two per cent of HAV positive cases required hospitalizations and had average hospitalization duration of 13.9 days, with the range of 7-30 days; one death (12 years female) was attributed to HAV.
Of the randomly picked 20 IgM ELISA positive samples, 16 were positive by nested RT PCR. Of these, 12 samples were subjected to sequencing and nine had satisfactory electropherogram data. BLAST analysis was used to confirm the identity. One representative sequence of each homologues group was submitted to Gen bank. The sequences of sample HEP-8 (Gen bank KC583366) HEP-52 (Gen bank KC583367) and HEP-208 (Gen bank KC583368) showed 100 per cent homology with Indian isolates having a Gen bank accession number FJ355790.1, FJ355896.1, FJ355612.1 respectively. The phylogenetic analysis revealed that the HAV virus circulating in and around Jabalpur belonged to genotype IIIA (Fig.).
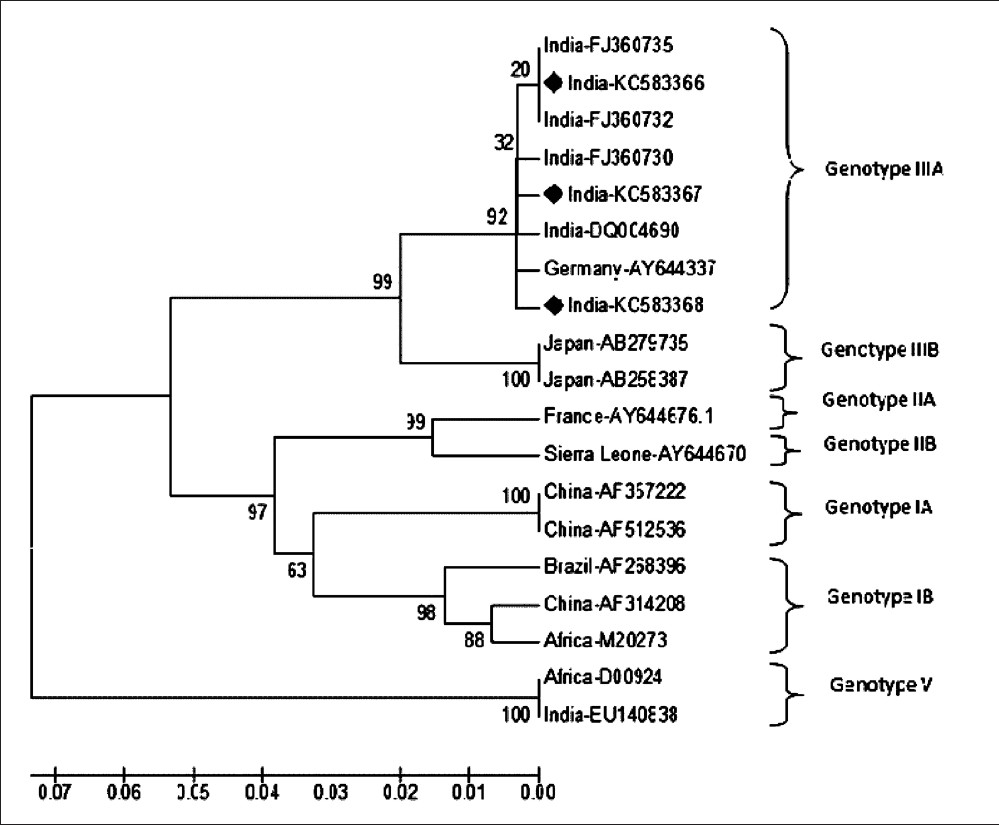
- Phylogenetic tree of hepatitis A viruses generated by the p distance NJ method, based on the 5’ NCR nucleotide sequence (292 bases). Each strain is labelled as country followed by Gene bank accession number. The strains from this study are marked with (♦). The respective genotypes are marked after bracket.
Among the HAV confirmed cases from urban area, 82 per cent (n=22) were from slum area where poor sanitation and unhygienic conditions prevailed. This number was higher than that reported earlier131415 probably because the samples in this study were referred from government hospitals, wherein primarily patients are from low socio-economic background are treated.
Outbreaks389 and higher number of sporadic cases16 of HAV have been reported in different seasons from India. In our study we observed an upsurge of cases in the rainy season as reported earlier16. Faecal contamination of water and inadequate water treatment are a major source of HAV infection1.
Of the three genotypes of HAV infecting humans, genotype IA, IB and IIIA are known to be circulating/co-circulating in different parts of India with predominance of genotype IIIA8910111517. The partial sequence analysis of 5’ NCR region indicated that genotype IIIA was the major genotype circulating in the central India. The severity of HAV infection is known to differ with genotype involved18 and different genotypes could be responsible for sporadic cases and outbreaks310. Thus isolation and molecular characterization to understand divergence of circulating HAV genotypes by targeting other regions of HAV genome such as VP1/2A junction, 2A, 2C and 3D for microanalysis are essential19.
Although our study has a limitation that it is based on the referred samples, yet adequate samples were analyzed and the study gives a preliminary epidemiological data. Longitudinal studies involving socio-economic status, hygienic conditions, access to safe drinking water with rigorous clinical monitoring supported with serological and molecular tools will help in understanding the intricacies of HAV epidemiology in central India.
The clinical and epidemiological picture of HAV infection is changing24. Moreover, it is difficult for the clinicians to differentiate between hepatitis viruses based on clinical and biochemical characteristics thus punctual and precise diagnosis would not only help in appropriate treatment and intervention, but also aid in curbing the possible outbreaks.
Acknowledgment
Authors thank the Secretary to Government of India, Department of Health Research (DHR), Ministry of Health & Family Welfare, and The Director-General, ICMR for financial support under ICMR's VDL network project. The authors also thank Director, RMRCT, Jabalpur for support and encouragement during the work and for critical review of the manuscript. Authors acknowledge Dr R. K. Sharma in statistical analysis of data and technical help by Shriyut M. J. Ukey and L. Sahare, staff of virology laboratory. The co-operation and support of clinicians of Seth Govind Das District Hospital and Netaji Subashchandra Bose Medical College and Hospital, Jabalpur, for sample collection and referral is acknowledged.
References
- WHO Media centre. Hepatitis A: Fact sheet N°328, updated. 2012. Available from: http://www.who.int/mediacentre/factsheets/fs328/en/
- [Google Scholar]
- Epidemiological transition of hepatitis A in India: issues for vaccination in developing countries. Indian J Med Res. 2008;128:699-704.
- [Google Scholar]
- Molecular characterization of hepatitis A virus from a large outbreak from Kerala, India. Indian J Med Res. 2006;123:760-9.
- [Google Scholar]
- Nalbantoglu A Shifting epidemiology of hepatitis a infection and vaccination status of children aged 6 months-12 years: time for mass vaccination. Iran J Pediatr. 2013;23:276-80.
- [Google Scholar]
- 2007. Hepatitis A virus. In: Knipe D, Howley P, eds. Fields virology (5th ed). Philadelphia, USA: Lippincott Williams Wilkins; 2007. p. :911-47.
- [Google Scholar]
- Characterization of the complete genomic sequence of genotype II hepatitis A virus (CF53/Berne isolate) J Gen Virol. 2004;85:2943-52.
- [Google Scholar]
- Genetic relatedness of hepatitis A virus strains recovered from different geographical regions. J Gen Virol. 1992;73:1365-77.
- [Google Scholar]
- Outbreaks of hepatitis A among children in western India. Trans R Soc Trop Med Hyg. 2009;103:911-6.
- [Google Scholar]
- Investigation of a hepatitis A outbreak from Shimla Himachal Pradesh. Indian J Med Res. 2009;130:179-84.
- [Google Scholar]
- Co-circulation of and co-infections with hepatitis A virus subgenotypes IIIA and IB in patients from Pune, western India. Hepatol Res. 2007;37:85-93.
- [Google Scholar]
- Increasing trend of acute hepatitis A in north India: need for identification of high-risk population for vaccination. J Gastroenterol Hepatol. 2006;21:689-93.
- [Google Scholar]
- MEGA 5: Molecular Evolutionary Genetics Analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28:2731-9.
- [Google Scholar]
- Exposure of Indian children to hepatitis A virus & vaccination age. Indian J Med Res. 1999;109:11-5.
- [Google Scholar]
- Hepatitis A in day-care centers. A community-wide assessment. N Engl J Med. 1980;302:1222-7.
- [Google Scholar]
- Changing epidemiology of hepatitis A and hepatitis E in urban and rural India (1982-98) J Viral Hepat. 2001;8:293-303.
- [Google Scholar]
- Acute sporadic viral hepatitis in urban population of tribal districts of Madhya Pradesh. Indian Pediatr. 1998;35:105-9.
- [Google Scholar]
- Full length genomes of genotype IIIA hepatitis A virus strains (1995-2008) from India and estimates of the evolutionary rates and ages. Infection, Genet Evol. 2009;9:1287-94.
- [Google Scholar]
- Comparative analysis of disease severity between genotypes IA and IIIA of hepatitis A virus. J Med Virol. 2011;83:1308-14.
- [Google Scholar]
- Evaluation of genomic regions of hepatitis A virus for phylogenetic analysis: SuitabilityAv of the 2C region for genotyping. J Virol Methods. 2008;153:36-42.
- [Google Scholar]






