Translate this page into:
TEM mediated extended spectrum cephalosporin resistance in clinical & environmental isolates of Gram negative bacilli: A report from northeast India
*For correspondence: srjoshi2006@yahoo.co.in
This is an open access article distributed under the terms of the Creative Commons Attribution NonCommercial ShareAlike 3.0 License, which allows others to remix, tweak, and build upon the work non commercially, as long as the author is credited and the new creations are licensed under the identical terms.
This article was originally published by Medknow Publications & Media Pvt Ltd and was migrated to Scientific Scholar after the change of Publisher.
Sir,
The increase in the number of extended spectrum beta lactamase (ESBL) producing Gram-negative bacilli (GNB) has always been a threat to healthcare. ESBL producing bacteria are commonly isolated from hospitalized patients but their dissemination into the environment especially water bodies has been a cause of concern1. Since ESBL genes are usually found encoded on mobile vectors such as plasmids (blaCTX-M, blaSHV) and transposons (blaTEM), the transfer of resistance between bacteria is easily facilitated23.
The present study was aimed to investigate the presence of ESBL producing bacteria belonging to Enterobacteriaceae in water bodies (rivers, ponds, lakes) and clinical setting of a high altitude city of north eastern India, Shillong, the capital city of Meghalaya.
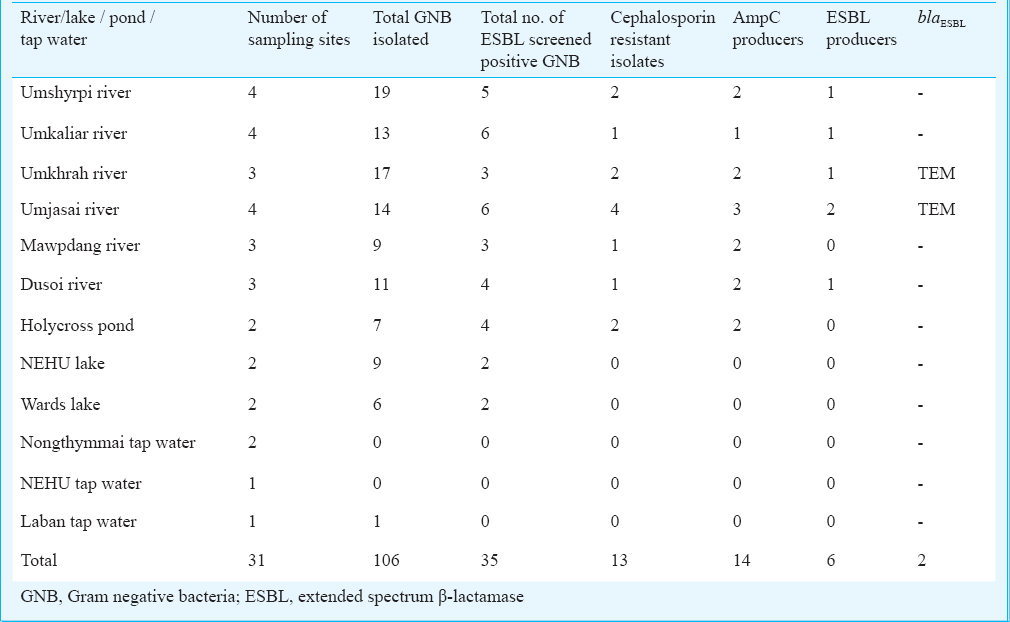
Water samples were collected in sterile vials from 31 water bodies including rivers, ponds, and tap water supply from Shillong, Meghalaya (Table I). For further analysis, serial dilutions were made starting with 1 ml of water samples diluted in 9 ml of saline solution (0.9% NaCl). A volume of 100 µl from each well homogenized dilution was inoculated onto the MacConkey agar plates (Hi-Media, Mumbai, India). Plates were incubated at 37°C for 24 h under aerobic conditions. All lactose fermenting and non-fermenting colonies with different colouration and morphology were picked from the selective plates, subcultured and stored in glycerol stock (2%) at -80°C.
To further compare the ESBL producers obtained from water samples with the clinical isolates, 10 consecutive, non-duplicate, cephalosporin resistant clinical isolates were taken from the Microbiology department of Nazareth Hospital and The Children's Hospital, Shillong. The study was carried out in the Microbiology laboratory of North Eastern Hill University (NEHU), Shillong, from August to December 2013.
All isolates were selected on the basis of their initial screening for presence of ESBL by combined disc diffusion method according to CLSI (Clinical and Laboratory Standards Institute) recommendation4. For amplification and characterization of blaESBL genes, a set of six primers was used namely: blaTEM, blaCTX-M, blaSHV, blaOXA-2, blaOXA-10 and blaGES as described elsewhere5. Reaction mixture was prepared using Promega PCR master mix (Promega, USA) and reaction were run under the following conditions: initial denaturation 94°C for 5 min, 33 cycles of 94 °C for 35 sec, 51°C for 1 min, 72°C for 1 min and final extension at 72°C for 7 min. The amplicons were sequenced and compared by performing BLAST analysis (http://blast.ncbi.nlm.nih.gov/Blast.cgi). Negative control used in the PCR reaction was Escherichia coli ATCC 25922, and three previously confirmed isolates from our laboratory of E. coli producing blaTEM, blaCTX-M, blaSHV were taken as positive control.
Screening for the presence of AmpC (class of C beta lactamases) and metallo beta-lactamases (MBL) was performed by cefoxitin disc test6 and imipenem EDTA combined disc test7, respectively. Multiplex PCR was performed for detection of MBL and AmpC gene types. PCR conditions and primers were as described previously89. For detection of class 1 and class 2 integron, integrase genes PCR were performed10. To find the genetic association of blaESBL gene with integrons, PCR was performed using forward primer of the conserved region (5’CS) of integron gene and reverse primer of the characterized ESBL gene as well as 3’CS region of integron and forward primer of ESBL gene8. Integrons containing functional genes known as gene cassettes, were amplified by 59 base elements PCR and amplicons were sequenced11.
Antimicrobial susceptibility testing was performed by Kirby Bauer disc diffusion method (CLSI, 2011)4 on Muller-Hinton agar plates using antibiotics viz. cefopodoxime (10 μg), amikacin (30 μg), gentamicin (10 μg), ciprofloxacin (30 μg), trimithoprim/sulphamethoxazole (1.25 / 23.75 μg), tigecycline (15 μg), cefepime (30 μg), imipenem (10 μg), meropenem (10 μg) and aztreonam (30 μg)4 (Hi-Media, Mumbai, India).
A total of four bacterial isolates belonging to Enterobacteriaceae family and non-fermenting rods were confirmed in this study as ESBL producers (Table II), two of which i.e. E. coli (R59) and Pseudomonas aeruginosa (R60) were from the environmental source (river water) and the remaining two were from clinical samples. Both the environmental isolates showed susceptibility towards imipenem, kanamycin, gentamicin, ciprofloxacin, tigecycline and polymixin B. Among the two ESBL producing isolates obtained from clinical samples, one KP7 (Klebsiella pneumoniae) was from the sputum of a one year old child admitted in intensive care unit, showed susceptibility to imipenem, ciprofloxacin, co-trimoxazole and polymyxin B while other isolate EC12 (E. coli) from the stool sample of a 16 yr old female was susceptible only towards polymyxin B.
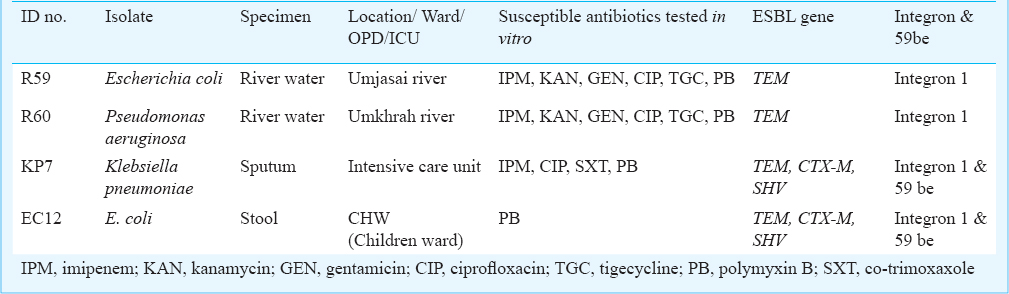
Multiplex PCR assay yielded the product with expected size for blaTEM for two of the environmental isolates (R59 & R60) from river flowing outside the hospital area and also from the clinical isolates. However, sequencing could not reveal the exact variant of TEM type ESBL. Chromosomal AmpC was found in all four isolates while none of these were carrying any MBL gene. Among TEM negative environmental isolates, AmpC production was suspected in 12 isolates, but none of these were harbouring any plasmid AmpC gene as targeted by multiplex PCR. Presence of TEM and class 1 integron was demonstrated in all the four isolates while TEM was found to be cassette mediated only in case of clinical isolates.
The two isolates obtained from water bodies showed susceptibility towards all the tested antibiotics except cephalosporins. However, the clinical isolate EC12 conferred resistance against quinolones and aminoglycosides, whereas KP7 was resistant to all the antibiotics except polymixin B.
The prevalence of ESBL in north-eastern part of India found to be varied from 67-74 per cent12. These resistance determinants detected in the characterized isolates may easily disseminate to the community during irrigation using river water1131415. Presence of these resistant genes within bacteria in river traces indicates their origin from anthropogenic sources, such as hospital, and municipal effluents16. It is quite alarming that this resistant determinant (TEM) is maintained within the organism in lotic water system where antibiotic pressure is minimum or absent.
In our country, most of the studies are focused on the beta lactamase gene obtained from isolates from hospital environment. However, this study underscores the persistence of the blaESBL along with gene capture mechanism in isolates from clinical and environmental sources. Maintenance of these resistant determinants in environmental reservoir system and their transmission to human host was evidenced by a recent study which showed that the healthy travellers acquired carbapenemases producing enterobacteriaceae upon their visit to India without any contact with local healthcare centres17. Thus, it may be suggested that appropriate measures are required to reduce the burden of antibiotic resistance in the environment as well as proper treatment of municipal and hospital waste water and improvement of water quality.
The present investigation was a pilot study, preliminary in nature and further research is necessary to determine the transferability of the resistant determinant or mechanism of gene transfer. Further, the presence of TEM mediated extended spectrum cephalosporin resistance would further restrict the therapeutic alternatives and infection control management strategies.
Acknowledgment
The authors acknowledge the Departments of Microbiology, Nazareth Hospital and The Children's Hospital, Shillong, for providing the clinical isolates. This work was sponsored by the University Grants Commission (UGC) under the Dr D.S. Kothari Postdoctoral Fellowship Scheme (No.F.4-2/2006(BSR)/13-927/2013(BSR) to the first author (SU).
References
- Characteristics of extended-spectrum beta-lactamase and carbapenemase-producing Enterobacteriaceae isolates from rivers and lakes in Switzerland. Appl Environ Microbiol. 2013;79:3021-6.
- [Google Scholar]
- Analysis of blaCTX-M carrying plasmids from Escherichia coli isolates collected in the BfT-GermVet study. Appl Environ Microbiol. 2011;77:7142-6.
- [Google Scholar]
- Distribution of the blaTEM gene and blaTEM-containing transposons in commensal Escherichia coli. J Antimicrob Chemother. 2011;66:745-51.
- [Google Scholar]
- Clinical and Laboratory Standards Institute (CLSI). Performance standards for antimicrobial susceptibility testing; 21 st Informational Supplement: M100-S21. Wayne, PA, USA: CLSI; 2011.
- [Google Scholar]
- Prevalence of Ambler class A and D â-lactamases among clinical isolates of Pseudomonas aeruginosa in Korea. J Antimicrob Chemother. 2005;56:122-7.
- [Google Scholar]
- Occurrence and detection of AmpC beta-lactamases among Escherichia coli, Klebsiella pneumoniae, and Proteus mirabilis isolates at a veterans medical center. J Clin Microbiol. 2000;38:1791-6.
- [Google Scholar]
- Imipenem-EDTA disk method for differentiation of metallo-b-lactamase-producing clinical isolates of Pseudomonas spp. and Acinetobacter spp. J Clin Microbiol. 2002;40:3798-801.
- [Google Scholar]
- Characterization of a new metallo-beta-lactamase gene, blaNDM-1, and a novel erythromycin esterase gene carried on a unique genetic structure in Klebsiella pneumoniae sequence type 14 from India. Antimicrob Agents Chemother. 2009;53:5046-54.
- [Google Scholar]
- Detection of plasmid-mediated AmpC â-lactamase genes in clinical isolates by using multiplex PCR. J Clin Microbiol. 2002;40:2153-62.
- [Google Scholar]
- Identification of epidemic strains of Acinetobacter baumannii by Integrase gene PCR. J Clin Microbiol. 2001;39:8-13.
- [Google Scholar]
- Gene cassette PCR: sequence-independent recovery of entire genes from environmental DNA. Appl Environ Microbiol. 2001;67:5240-6.
- [Google Scholar]
- Prevalence of blaTEM, blaSHV and blaCTX-M genes in clinical isolates of Escherichia coli and Klebsiella pneumoniae from Northeast India. Indian J Pathol Microbiol. 2014;57:249-54.
- [Google Scholar]
- Isolation of extended spectrum â-lactamase (ESBL) producing bacteria from urban surface waters in Malaysia. Malays J Med Sci. 2013;20:14-22.
- [Google Scholar]
- The prevalence of extended-spectrum â-lactamase in environmental isolates of Enterobacter. Indian J Pathol Microbiol. 2008;51:130-6.
- [Google Scholar]
- High diversity of extended-spectrum beta-lactamase producing bacteria in an urban river sediment habitat. Appl Environ Microbiol. 2010;76:5972-6.
- [Google Scholar]
- Detection of extended-spectrum beta-lactamase-producing Klebsiella pneumoniae in effluents and sludge of a hospital sewage treatment plant. Lett Appl Microbiol. 2008;46:136-41.
- [Google Scholar]
- Acquisition of carbapenemase- producing Enterobacteriaceae by healthy travellers to India, France, February 2012 to March 2013. Euro Surveill. 2014;19 pii: 20768
- [Google Scholar]





