Translate this page into:
Phylogenetic analysis of swine & human hepatitis E virus
*For correspondence: shambhavi.srivastava@gmail.com
This is an open-access article distributed under the terms of the Creative Commons Attribution-Noncommercial-Share Alike 3.0 Unported, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
This article was originally published by Medknow Publications and was migrated to Scientific Scholar after the change of Publisher.
Sir,
We have read with interest a recent paper published in the journal1. The authors amplified genomic material from swine sera using primers based on hepatitis E virus (HEV) genomic sequences, and sequenced the amplicons so obtained. They drew a phylogenetic tree based on the sequences of these Indian swine isolates, and genomic sequences of HEV isolates from humans and animals.
The PCR amplicons which were 246-nucleotide long, included sequences of the primers used for PCR. The primer part of the amplicon is provided from outside. Hence it may not accurately represent the sequence of the target nucleic acid and cannot be used for sequence analysis. Thus, for phylogenetic analysis, only the middle part of the sequence after removing the primer sequences on either side should be used. Removal of the primers in this case would have left a 196-nucleotide long sequence (i.e. 246-nucleotide PCR product minus 22 and 28-nucleotide forward and reverse primers); this would have been the appropriate region for comparison with other sequences.
We have redrawn a phylogenetic tree using only this 196-nucleotide segment after omitting the primer regions from the sequence used. This tree (Fig.) is though similar to that drawn by the authors1 shows minor variations in the branching pattern of the branch lengths in same clades. However, we thought that we will take this opportunity to correct this minor error in the published record.
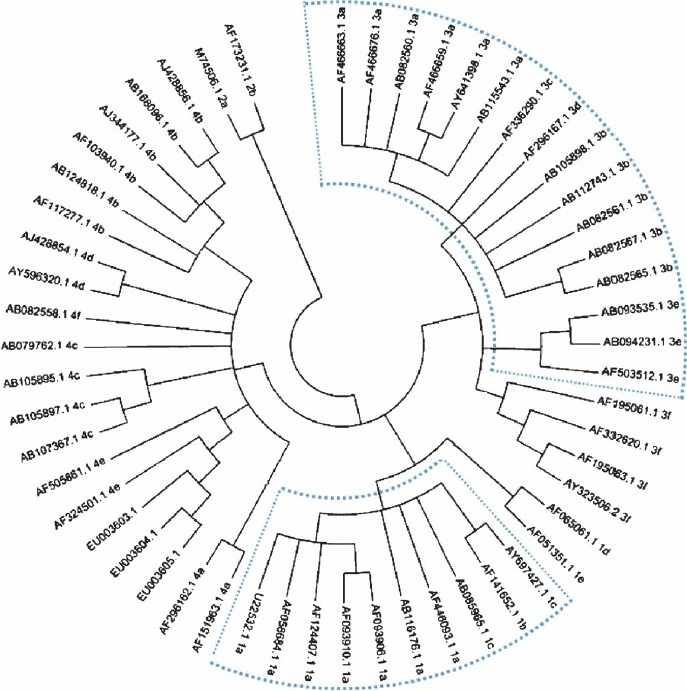
- Phylogenetic relationship among swine and human isolates of hepatitis E virus representing the 4 major genotypes, based on a 196-nucleotide fragment of ORF2 of the viral genome. The regions of the tree showing differences from the published tree are highlighted using dotted lines.
References
- Molecular analysis of swine hepatitis E virus from north India. Indian J Med Res. 2010;132:504-8.
- [Google Scholar]





