Translate this page into:
Optimization of loop-mediated isothermal amplification-based method for detection of macrolide–lincosamide–streptogramin B resistance in Staphylococcus aureus
* For correspondence: ab0404@gmail.com
-
Received: ,
This article was originally published by Wolters Kluwer - Medknow and was migrated to Scientific Scholar after the change of Publisher.
Sir,
The growing interest in the resistance of Staphylococcus aureus to various antibiotics in the last two decades has led to re-considerations around usage of macrolide–lincosamide–streptogramin B (MLSb) antibiotics as erythromycin and clindamycin since long have been potent options for treating both methicillin-resistant S. aureus (MRSA) and methicillin-susceptible S. aureus (MSSA). Unfortunately, in due course of time, resistance to MLSb antibiotics emerged in staphylococci and spread to different regions1-3. The common mechanisms of resistance to MLSb antibiotics are efflux of antibiotics by the msrA gene, target site modification mediated by erm genes and inactivation of macrolides mediated by mphC genes4. To date, detection of MLSb resistance in S. aureus has relied on traditional phenotypic methods such as agar dilution technique, double-disc diffusion test and PCR-based molecular methods1-3,5. Although these methods exist for detection, these are time-consuming and require sophisticated laboratory equipment and reagents as well as ample technical expertise preventing the use of these techniques in low resource settings. This study was performed to explore the utility of a recently developed alternative technique called loop-mediated isothermal amplification (LAMP). It is a novel single-step technique where four to six sets of primers bind to distinct regions of the target DNA6. This assay has higher sensitivity and specificity in comparison to the conventional methods and allows users with low resources to avoid expensive equipment, reagents and tedious protocols7-9.
In this study, of the 168 clinical isolates of S. aureus, 40, which showed resistance to different MLSb antibiotics, were included. The MLSb-resistant phenotypes were determined by D-zone/disc diffusion test as per CLSI 2017 guidelines10. S. aureus 25923 was used as a control strain. Of the 40 isolates, exhibiting MLSb resistance, 20 (50%) expressed constitutive MLSb (cMLSb) phenotype, seven (17.5%) isolates expressed inducible MLSb (iMLSb) phenotype and 13 (32.5%) expressed MSb phenotype. Total DNA from all the bacterial strains was extracted by boiling centrifugation method - to be used as a template. A conventional polymerase chain reaction (PCR) was performed with oligonucleotide primers (Supplementary Table I) targeting the MLSb resistance genes ermA, ermB, ermC, lnu, msrA and mphC. Each single reaction mixture (25 µl) contained 2 µl of template DNA (100 ng/µl), 1 µl each of primer (10 picomoles), 12.5 µl GoTaq Green Master Mix 2X DNA Polymerase (Promega, Madison, USA) and molecular grade nuclease free water. The PCR reactions were performed in a thermal cycler with 35 cycles of initiation, annealing and extension. The PCR assay revealed that of the 40 isolates, 34 (85%) harboured msrA and mphC genes either alone or in combination. No other MLSb resistance genes could be detected in the remaining six (15%) isolates. The details of the MLSb resistance profile of all the 40 isolates are given in Supplementary Table II.
| Primer name | Primer pairs | Product length (bp) | Reference |
|---|---|---|---|
| mphC (F) mphC (R) | 5’- ACTTACAGGCAAACCCGCAG-3’ 5’- GTCCATTGACGGATCGGAGT-3’ |
412 | Hauschild and Schwarz 201011 |
| msr A (F) msr A (R) | 5’- TCCAATCATTGCACAAAATC-3’ 5’- AATTCCCTCTATTTGGTGGT-3’ |
163 | Duran et al., 201212 |
| erm A (F) erm A (R) | 5’- AAGCGGTAAACCCCTCTGA-3’ 5’- TTCGCAAATCCCTTCTCAAC-3’ |
190 | Duran et al., 201212 |
| erm B (F) erm B (R) | 5’- CTATCTGATTGTTGAAGAAGGATT-3’ 5’- GTTTACTCTTGGTTTAGGATGAAA-3’ |
142 | Duran et al., 201212 |
| erm C (F) erm C (R) | 5’- AATCGTCAATTCCTGCATGT-3’ 5’- TAATCGTGGAATACGGGTTTG-3’ |
299 | Duran et al., 201212 |
| vga ( F) vga ( R) | 5’- CGCCATCTGTCAAAATCGGT-3’ 5’- AACTCGCTCTCCACCACTTA-3’ |
191 | Allignet et al., 199313 |
| lnu B (F) lnu B (R) | 5’- GATGTACGACGCACCAAACG-3’ 5’- CCAGTTCTTGGCGGTAAGGT-3’ |
345 | Si et al., 201514 |
| Isolate ID | MLSb resistance phenotypes as detected by D-zone/double-disc diffusion test | Resistance profile | Detection by PCR (msrA and mphC) |
|---|---|---|---|
| CS1 | cMLSb | ERY-R, CLI-R | msrA |
| CS8 | cMLSb | ERY-R, CLI-R | msrA |
| CS4 | iMLSb | ERY-R, CLI-S (D-shaped zone of inhibition) | msrA |
| CS14 | iMLSb | ERY-R, CLI-S (D-shaped zone of inhibition) | msrA |
| CS17 | MSb | ERY-R, CLI-S | msrA |
| CS15 | MSb | ERY-R, CLI-S | msrA |
| CS21 | MSb | ERY-R, CLI-S | msrA |
| CS23 | cMLSb | ERY-R, CLI-R | msrA |
| CS22 | cMLSb | ERY-R, CLI-R | msrA |
| CS26 | MSb | ERY-R, CLI-S | msrA |
| CS27 | iMLSb | ERY-R, CLI-S (D-shaped zone of inhibition) | msrA |
| CS30 | iMLSb | ERY-R, CLI-S (D-shaped zone of inhibition) | msrA |
| CS32 | cMLSb | ERY-R, CLI-R | msrA |
| CS35 | cMLSb | ERY-R, CLI-R | msrA |
| CS37 | cMLSb | ERY-R, CLI-R | msrA |
| CS39 | iMLSb | ERY-R, CLI-S (D-shaped zone of inhibition) | msrA |
| CS2 | cMLSb | ERY-R, CLI-R | msrA, mphC |
| CS5 | cMLSb | ERY-R, CLI-R | msrA, mphC |
| CS6 | iMLSb | ERY-R, CLI-S (D-shaped zone of inhibition) | msrA, mphC |
| CS9 | cMLSb | ERY-R, CLI-R | msrA, mphC |
| CS10 | cMLSb | ERY-R, CLI-R | msrA, mphC |
| CS12 | MSb | ERY-R, CLI-S | msrA, mphC |
| CS16 | cMLSb | ERY-R, CLI-R | msrA, mphC |
| CS19 | MSb | ERY-R, CLI-S | msrA, mphC |
| CS24 | cMLSb | ERY-R, CLI-R | msrA, mphC |
| CS25 | cMLSb | ERY-R, CLI-R | msrA, mphC |
| CS28 | cMLSb | ERY-R, CLI-R | msrA, mphC |
| CS29 | MSb | ERY-R, CLI-S | msrA, mphC |
| CS31 | cMLSb | ERY-R, CLI-R | msrA, mphC |
| CS33 | cMLSb | ERY-R, CLI-R | msrA, mphC |
| CS34 | MSb | ERY-R, CLI-S | msrA, mphC |
| CS36 | cMLSb | ERY-R, CLI-R | msrA, mphC |
| CS38 | MSb | ERY-R, CLI-S | msrA, mphC |
| CS40 | MSb | ERY-R, CLI-S | msrA, mphC |
| CS3 | MSb | ERY-R, CLI-S | Negative |
| CS7 | MSb | ERY-R, CLI-S | Negative |
| CS11 | cMLSb | ERY-R, CLI-R | Negative |
| CS13 | MSb | ERY-R, CLI-S | Negative |
| CS18 | iMLSb | ERY-R, CLI-S (D-shaped zone of inhibition) | Negative |
| CS20 | cMLSb | ERY-R, CLI-R | Negative |
*ERY-R, erythromycin resistant; *CLI-R, clindamycin resistant; *CLI-S, clindamycin susceptible; MLSb, macrolide-lincosamide-streptogramin B; cMLSb, constitutive MLSb; iMLSb, inducible MLSb; PCR, polymerase chain reaction; MSb, macrolide-streptograminb
Sequences of msrA (Accession No. KX211999) and mphC (Accession No. GQ183071) genes were retrieved from NCBI nucleotide database. Six sets of primers, namely two inner primers (FIP and BIP), two outer primers (F3 and B3) and two loop primers (LFP and BFP) targeting six distinct regions, were designed for each of these genes using LAMP PrimerExplorer V4 (http://prjjjimerexplorer.jp). The sequences of the designed LAMP primers are listed in Table. The positions of the LAMP primers in msrA and mphC gene fragment are given in Supplementary Figures
| msrA | ||
|---|---|---|
| Primer | Sequence (5’- 3’) | Length |
| FIP (F1c+F2) | ACGAGCGCTATATTTTTGCCATAT-GAAGTCAAAAACTGCTAACACA | 46 |
| BIP (B1c+B2) | TACCACCAAATAGAGGGAATTGATT-TTCATAAGCAAGTTGACGATAG | 47 |
| F3 | ATTGCACAAAATCTAACATTGG | 22 |
| B3 | TGAAACGTCACGCATGTC | 18 |
| LFP | GGTATTTGGAATCGTAC | 17 |
| LBP | GTTCTCCTAAAGTGC | 15 |
| mphC | ||
| Primer | Sequence (5’- 3’) | Length |
| FIP (F1c+F2) | TGGATGTAAGTCTCCATGTATCATG-AATGGTTAGAAAACGACGAACT | 47 |
| BIP (B1c+B2) | TTACCAAGCAAATGTCATAGGACT-TCCATTGACGGATCGGAG | 42 |
| F3 | GGATTATGGAACAGATGGAAAC | 22 |
| B3 | AATACACGATGGTATCCCAT | 20 |
| LFP | TTGCACGTCGAGGCCAC | 17 |
| LBP | AGACTGGACTGAAGCAACCT | 20 |
FIP, forward inner primer; BIP, backward inner primer; LFP, loop forward primer; LBP, loop backward primer
The LAMP reaction mixture was optimized by modifying the components of the reaction mixtures, reaction duration (30-60 min) and incubation temperature (55-65ºC). The 25 µl optimized reaction mixture contained 2.5 µl 10X ThermoPol buffer (NEB, England), 1.5 µl MgSO4 (NEB, England), 3.5 µl dNTP mix (1.4 mM each, HiMedia, India), 4 µl of primer mix (2 µM FIP, 2 µM BIP, 1.2 µM F3, 1.2 µM B3, 1.6 µM LF and 1.6 µM BF), 1 µl Bst DNA polymerase (NEB, England) and 2 µl (100 ng) template. To optimize reaction temperature, the reaction mixture was incubated at different temperatures. The temperatures were increased from 55 to 65ºC, i.e. 55ºC, 57ºC, 62ºC and 65ºC, for different lengths of time, i.e. 30, 40, 50 and 60 min, followed by heating to 80ºC for 10 min for enzyme inactivation to stop the reaction. The incubation was carried out in a thermal cycler. S. aureus ATCC 25,923 without any MLSb resistance genes was used as a negative control. A no template control containing 5 µl sterile water was also simultaneously run. Various modes are available for detection of LAMP products. Naked-eye detection by addition of metal indicators, fluorescence detection with intercalating agents, lateral flow and agarose gel electrophoresis - all are compatible with LAMP reactions. In this study, the LAMP products were electrophoresed in two per cent agarose gel and visualized in an ultraviolet light transilluminator in a Gel Doc EZ imager (Bio-Rad, USA) to confirm the amplification. The presence of ladder-like bands is the typical electrophoresis pattern of amplification by LAMP reaction15. The optimum reaction temperature and duration were found to be 65ºC and 60 min, respectively, as the best intensity ladder-like pattern was observed in this reaction condition (Fig. A and B). All the 40 clinical isolates were assessed by LAMP assay. It was observed that the optimized LAMP assay was able to detect the presence of msrA and mphC genes in all 34 isolates as was detected with conventional PCR assay. No non-specific reaction was observed in the remaining six isolates as well as in S. aureus ATCC 25923 isolate, which tested negative by PCR.
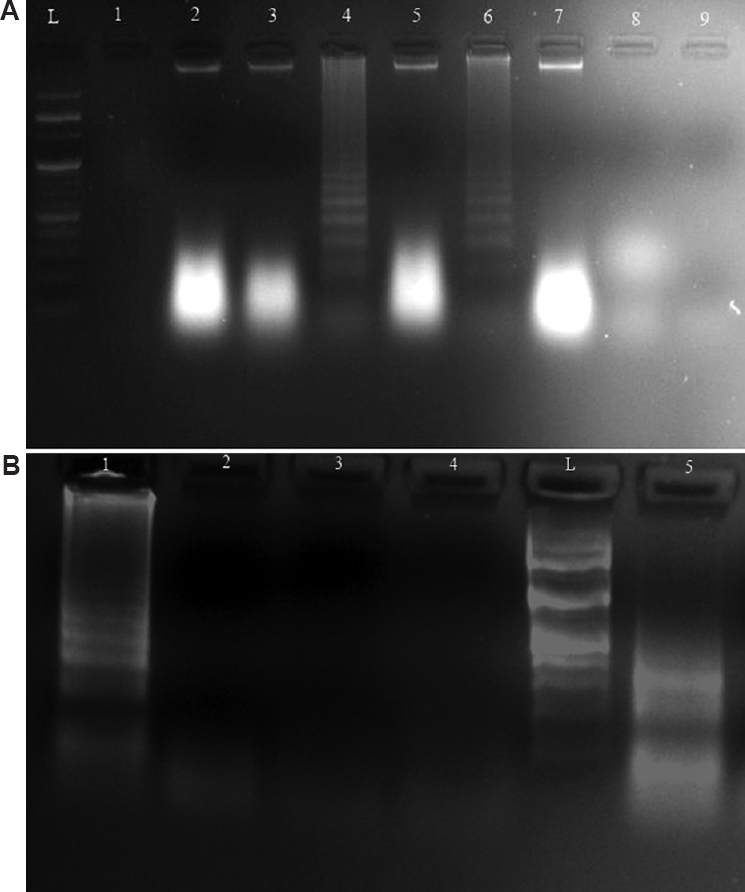
- (A) Electrophoretic analysis of LAMP-amplified msrA gene in S. aureus at 65, 62 and 57ºC. (A) Lane L, 100 bp DNA ladder; Lanes 4 and 6, Characteristic ladder pattern at 65ºC; Lanes 2 and 5, Faint smear at 62ºC; Lanes 3 and 7, Faint smear at 57ºC; Lane 8, No characteristic pattern in E. coli; Lane 9, no template control; (B) Electrophoretic analysis of LAMP-amplified mphC gene in S. aureus at 65, 62 and 57ºC. Lane L, 100 bp DNA ladder; Lane 1, Characteristic ladder pattern at 65ºC; Lane 5, Ladder-like pattern at 62ºC; Lane 4, no characteristic pattern at 57ºC; Lane 3, No characteristic pattern in E. coli; Lane 2, no template control.
MLSb antibiotics have been potent options for treating both MRSA and MSSA in India since long. Acquired genes such as msrA and mphC, which code for ATP-dependent efflux pumps and phosphotransferases, respectively, confer strong resistance to 14- and 15-membered macrolides and streptogramin B16. Due to high prevalence of MLSb resistance genes, early detection and timely clinical intervention is necessary to contain their further spread. Although conventional phenotypic and molecular detection techniques such as D-zone test and PCR are available, these are time-consuming and resource intensive. Moreover, in phenotypic tests such as D-zone test, the results are often affected owing to numerous factors such as inoculum size, formulation of media, rate of growth, incubation condition and duration17,18. In conventional nucleic acid-based detection methods such as PCR, the use of sophisticated instruments, high cost of reagents and the employment of highly qualified personnel for handling, are the main constraints in low-resource laboratories. Moreover, conventional PCR requires 35 cycles or more for the production of large quantity of amplicons which could increase the chances of generation of undesirable secondary amplicons19,20. It is, therefore, important for the routine microbiology laboratories to have access to a user-friendly method for rapid identification of resistant strains, which will facilitate early clinical intervention.
With this need in view, the present study optimized a LAMP assay for the purpose of detecting MLSb resistance genes msrA and mphC in clinical isolates of S. aureus. In this study, at an optimum temperature of 65ºC, the LAMP assay could yield result in 60 min of incubation. This is one of the outstanding features of LAMP where the amplification proceeds at a constant temperature by Bst exopolymerase having high strand displacement activity6. This effectively eliminates the need for tedious optimization of cycling conditions as required in PCR. The results of the LAMP assay were compared with that of the conventional PCR, and it was observed that the LAMP results were consistent with that of traditional PCR based analysis. No non-specific reaction with PCR-negative isolates was observed. This shows very high specificity of the LAMP assay while accurately detecting the genes msrA and mphC. Other studies have also shown that the LAMP assay is a good alternative to conventional PCR-based methods for its specificity and uniform temperature requirements, making it more convenient for microbiology laboratories to perform on a routine basis8,20. The specificity is generally high because the assay uses four sets of primers, which identify a number of distinct locations in the target DNA, thus eliminating the chances of primer mismatch, which often occurs in PCR. Our study came up with four sets of specific primers for each gene, which recognized six distinct regions. In addition to the inner and outer primers, loop primers were also designed, which identified distinct locations in each of the genes, and characteristic ladder pattern bands demonstrated the efficacy of the designed primers. LAMP products can also be easily visualized with naked eye in resource-poor areas where agarose gel electrophoresis and a gel documentation system are unavailable. Colorimetric detection with the addition of metal indicators and fluorescent detection with the addition of intercalating agents are some of the simple detection techniques, which are feasible in low-resource settings. However, cost evaluation and impact on therapeutic intervention should be carried out before considering its routine implementation in clinical microbiology laboratories. Another important feature of LAMP assay is its short operation time. The optimized LAMP assay in the present investigation had an operating time of 60 min as opposed to 90 min of the conventional PCR assay. Since the reaction proceeds at a constant temperature, the time loss due to temperature changes at different stages of amplification in conventional PCR is prevented21. The advantage of LAMP is not only in saving time but also due to its use of six sets of specific primers, which recognize distinct regions of the gene, making it more specific, and hence reducing the chances of non-specific amplification and false-positive reactions as compared to the conventional PCR. Short operation time and the use of six sets of specific primers thus give the LAMP-assay an edge over conventional PCR.
In conclusion, because MLSb resistance in S. aureus is an emerging threat and timely diagnosis and appropriate use of antibiotics is required, the LAMP assay optimized in this study has the potential to be adapted in any microbiology laboratory. This will also help in estimating resistance burden and informing appropriate therapy at a larger scale.
Financial support and sponsorship
The authors received financial support from Science and Engineering Research Board, Department of Science and Technology, Government of India vide no. (DST No.: EMR/2016/0055226).
Conflicts of interest
None.
Supplementary Fig. 1
Supplementary Fig. 1 Location of the primer sequences used in LAMP assay. The positions of the LAMP primers of msrA gene fragment of Staphylococcus aureus (Accession No. KX211999) are shown. Left and right arrows show complementary and sense sequences. F3 and B3 are outer primers; FIP (F1c + F2) is forward inner primer; BIP (B1c + B2) is backward inner primer.Supplementary Fig. 2
Supplementary Fig. 2 Location of the primer sequences used in LAMP assay. The positions of the LAMP primers of mphC gene fragment of Staphylococcus aureus (Accession No. GQ183071) are shown. Left and right arrows show complementary and sense sequences. F3 and B3 are outer primers; FIP (F1c + F2) is forward inner primer; BIP (B1c + B2) is backward inner primer.Acknowledgment
The authors acknowledge Biotech Hub, Assam University, Silchar, India, for providing the infrastructure
References
- Detection of macrolide and disinfectant resistance genes in clinical Staphylococcus aureus and coagulase-negative staphylococci. BMC Res Notes. 2011;4:453.
- [Google Scholar]
- Prevalence of inducible clindamycin resistance in Staphylococcus aureus isolated from clinical samples. Med J Armed Forces India. 2014;70:43-7.
- [Google Scholar]
- Prevalence of genotypes that determine resistance of staphylococci to macrolides and lincosamides in serbia. Front Public Health. 2017;5:200.
- [Google Scholar]
- Nomenclature for macrolide and macrolide-lincosamide-streptogramin B resistance determinants. Antimicrob Agents Chemother. 1999;43:2823-30.
- [Google Scholar]
- Detection and prevalence of inducible clindamycin resistance in staphylococci. J Med Microbiol. 2007;56:342-5.
- [Google Scholar]
- Loop-mediated isothermal amplification (LAMP): A rapid, accurate, and cost-effective diagnostic method for infectious diseases. J Infect Chemother. 2009;15:62-9.
- [Google Scholar]
- Sensitive and rapid detection of Clonorchis sinensis infection in fish by loop-mediated isothermal amplification (LAMP) Parasitol Res. 2010;106:1379-83.
- [Google Scholar]
- Comparative evaluation of the LAMP assay and PCR-based assays for the rapid detection of Alternaria solani. Front Microbiol. 2018;9:2089.
- [Google Scholar]
- Application of a loop-mediated isothermal amplification (LAMP) assay targeting cox1 gene for the detection of Clonorchis sinensis in human fecal samples. PLoS Negl Trop Dis. 2017;11:e0005995.
- [Google Scholar]
- Performance standards for antimicrobial susceptibility testing;twenty-first informational supplements, M100-S27. Wayne, PA: Clinical and Laboratory Standards Institute; 2017.
- Macrolide resistance in Staphylococcus spp. from free-living small mammals. Vet Microbiol. 2010;144:530-1.
- [Google Scholar]
- Antibiotic resistance genes &susceptibility patterns in staphylococci. Indian J Med Res. 2012;135:389-96.
- [Google Scholar]
- Sequence of a staphylococcal gene, vat, encoding an acetyltransferase inactivating the A-type compounds of virginiamycin-like antibiotics. Gene. 1993;130:91-8.
- [Google Scholar]
- Novel plasmid-borne multidrug resistance gene cluster including lsa(E) from a linezolid-resistant Enterococcus faecium isolate of swine origin. Antimicrob Agents Chemother. 2015;59:7113-6.
- [Google Scholar]
- Loop-mediated isothermal amplification (LAMP) of gene sequences and simple visual detection of products. Nat Protoc. 2008;3:877-82.
- [Google Scholar]
- The prevalence of genotypes that determine resistance to macrolides, lincosamides, and streptogramins B compared with spiramycin susceptibility among erythromycin-resistant Staphylococcus epidermidis . Mem Inst Oswaldo Cruz. 2016;111:155-60.
- [Google Scholar]
- D-Zone test for detection of inducible clindamycin resistance using SirScan paper disks and Rosco Neo-Sensitabs at 25 and 15 mm distances. J Med Microbiol. 2014;63:1052-4.
- [Google Scholar]
- Current and emerging methods of antibiotic susceptibility testing. Diagnostics (Basel). 2019;9:49.
- [Google Scholar]
- Emerging technologies for the clinical microbiology laboratory. Clin Microbiol Rev. 2014;27:783-822.
- [Google Scholar]
- Loop-mediated isothermal amplification (LAMP) reaction as viable PCR substitute for diagnostic applications: A comparative analysis study of LAMP, conventional PCR, nested PCR (nPCR) and real-time PCR (qPCR) based on Entamoeba histolytica DNA derived from faecal sample. BMC Biotechnol. 2020;20:34.
- [Google Scholar]
- Loop-mediated isothermal amplification (LAMP): A versatile technique for detection of micro-organisms. J Appl Microbiol. 2018;124:626-43.
- [Google Scholar]





