Translate this page into:
Multiplex polymerase chain reaction of genetic markers for detection of potentially pathogenic environmental Legionella pneumophila isolates
Reprint requests: Dr Rama Chaudhry, 2059, II Floor, Teaching Block, Department of Microbiology, All India Institute of Medical Sciences, New Delhi 110 029, India e-mail: drramach@gmail.com
-
Received: ,
This is an open access article distributed under the terms of the Creative Commons Attribution-NonCommercial-ShareAlike 3.0 License, which allows others to remix, tweak, and build upon the work non-commercially, as long as the author is credited and the new creations are licensed under the identical terms.
This article was originally published by Medknow Publications & Media Pvt Ltd and was migrated to Scientific Scholar after the change of Publisher.
Abstract
Background & objectives:
Genomic constitution of the bacterium Legionella pneumophila plays an important role in providing them a pathogenic potential. Here, we report the standardization and application of multiplex polymerase chain reaction (PCR) for the detection of molecular markers of pathogenic potential in L. pneumophila in hospital environment.
Methods:
Culture of the standard strains of L. pneumophila was performed in buffered charcoal-yeast extract agar with L-cysteine at pH 6.9. Primers were designed for multiplex PCR, and standardization for the detection of five markers annotated to L. pneumophila plasmid pLPP (11A2), lipopolysaccharide synthesis (19H4), CMP-N-acetylneuraminic acid synthetase (10B12), conjugative coupling factor (24B1) and hypothetical protein (8D6) was done. A total of 195 water samples and 200 swabs were collected from the hospital environment. The bacterium was isolated from the hospital environment by culture and confirmed by 16S rRNA gene PCR and restriction enzyme analysis. A total of 45 L. pneumophila isolates were studied using the standardized multiplex PCR.
Results:
The PCR was sensitive to detect 0.1 ng/μl DNA and specific for the two standard strains used in the study. Of the 45 hospital isolates tested, 11 isolates had four markers, 12 isolates had three markers, 10 isolates had two markers, nine isolates had one marker and three isolates had none of the markers. None of the isolates had all the five markers.
Interpretation & conclusions:
The findings of this study showed the presence of gene markers of pathogenic potential of the bacterium L. pneumophila. However, the genomic constitution of the environmental isolates should be correlated with clinical isolates to prove their pathogenic potential. Rapid diagnostic methods such as multiplex PCR reported here, for elucidating gene markers, could help in future epidemiological studies of bacterium L. pneumophila.
Keywords
Hospital infection control
Legionella pneumophila
multiplex polymerase chain reaction
The bacterium Legionella pneumophila, responsible for 90 per cent cases of Legionnaires’ disease, is found universally in various natural and artificial aquatic environments, including hospital water distribution systems12. Along with the host-associated risk factors, the genetic constitution of L. pneumophila plays a crucial role in establishing an infection345. As Legionellosis is generally considered a preventable infection, identifying and treating the aquatic sources, especially in hospitals, could help in the context of preventing hospital-acquired infection67. In 2010, a group from the Netherlands reported five gene markers highly correlated with clinical strains of L. pneumophila, annotated to L. pneumophila plasmid pLPP (11A2), lipopolysaccharide synthesis (19H4), CMP-N-acetylneuraminic acid synthetase (10B12), conjugative coupling factor (24B1) and hypothetical protein (8D6)8. This study was undertaken with the objective of finding L. pneumophila in our hospital environment and to know the distribution of genes which are considered to have pathogenic potential. We report here the development of a multiplex polymerase chain reaction (PCR) for the detection of these five gene markers and its application in our hospital environmental isolates of L. pneumophila.
Material & Methods
This study was conducted in the department of Microbiology, All India Institute of Medical Sciences, New Delhi, India during 2012-2015. The study was approved by the Institutional Bio-Safety Committee (water and swab samples - 200 each approved for collection). No human subjects and/or animals were involved in this study.
Culture and DNA extraction: The standard strains ATCC 33152 L. pneumophila strain Philadelphia-1 and ATCC 33153 L. pneumophila strain Knoxville were grown on buffered charcoal-yeast extract agar (BD-BBL, France) supplemented with 4 per cent L-cysteine-HCl (HiMedia laboratories, Mumbai) in a candle jar incubated at 37°C for three days. Gram stain was done and blood agar was used as a negative control plate. DNA was extracted from the colonies by the boiling method. Briefly, colonies were boiled in sterile distilled water for 10 min and centrifuged at 10,000 ×g to get the supernatant having the DNA.
PCR analysis and restriction enzyme analysis (REA): PCR detection of the 16S rRNA gene and REA were used for identification and confirmation of L. pneumophila standard strains and environmental isolates9. The restriction enzyme TaaI (HpyCH4III endonuclease, Thermo-Scientific Ltd., USA) was used for the differentiation of L. pneumophila and non-pneumophila Legionella species. Briefly, 15 μl of PCR product was digested with ×10 buffer Tango, 1U TaaI enzyme and adjusted with sterile water to final volume of 60 μl. The reaction was carried out in a water bath at 65°C for one hour and 20 μl of digested products were checked using 1.5 per cent agarose gel electrophoresis.
Multiplex PCR, sensitivity and specificity: Standardization of multiplex PCR for detection of five markers annotated to L. pneumophila plasmid pLPP (11A2), lipopolysaccharide synthesis (19H4), CMP-N-acetylneuraminic acid synthetase (10B12), conjugative coupling factor (24B1) and hypothetical protein (8D6) was done using the available gene sequences (Acc.nos. HM584933-HM584937). Primers (Table I) were selected and designed using NCBI Primer BLAST programme, and MPprimer design software10 was used for multiplex PCR primer design. The primers were synthesized commercially (Sigma, USA) and the DNA from both the standard strains were used for the multiplex PCR. Briefly, each reaction mix contained 5 μl of DNA, 5 μl of ×10 PCR buffer (Bangalore Genei, India), 1.5 μl of 10 mmol dNTP mix (Thermo Scientific), 0.5 μl of 10 pmol/μl each of five sets of primer, 0.5 μl of 3 U/μl Taq polymerase (Bangalore Genei) and sterile water adjusted to a final volume of 50 μl. The reaction was carried out in a thermal cycler (Applied Biosystems, USA) with the thermal programme having initial activation of 94°C for five minutes, 30 cycles of 94°C, 56°C, 72°C for 30 sec each and final extension of 72°C for 10 min. The primer pairs were checked individually for their sensitivity using various dilutions of standard strains’ DNA and specificity with DNA isolated from various bacteria. The individual PCR products were purified using QIAquick Gel extraction kit (Qiagen, Germany) and sequenced commercially to confirm their identity.
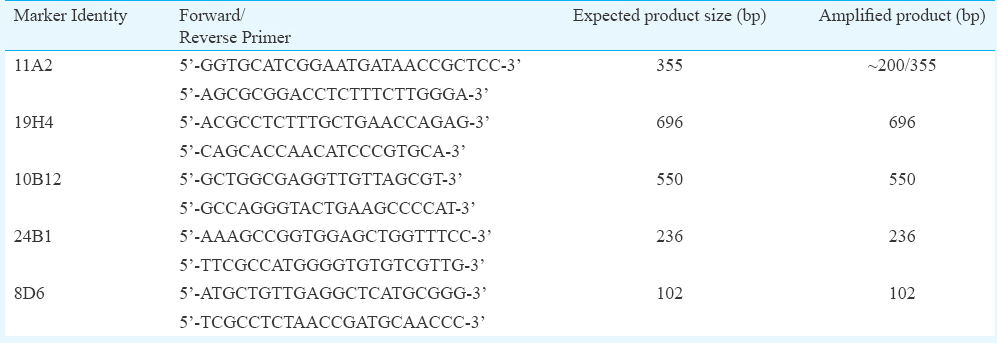
Hospital environmental sample collection and processing: A total of 145 water samples (both potable drinking water and non-potable water used for bathing, washing, gardening, cooling tower water) and 200 swab samples (pre-water collection) were collected and processed from July 2012 to May 2014. The samples were collected from distal outlets and from AC cooling towers (basin beneath the tower). Temperatures of the samples during the collection were also noted using a mercury thermometer (Zeal, England). The samples were treated with 0.1 N sodium thiosulphate and filtered with 0.22 μm mixed cellulose ester filters (Millipore, Germany). The filtrate was subjected to acid and thermal treatment and processed further for bacteriologic examination11 (Fig. 1).
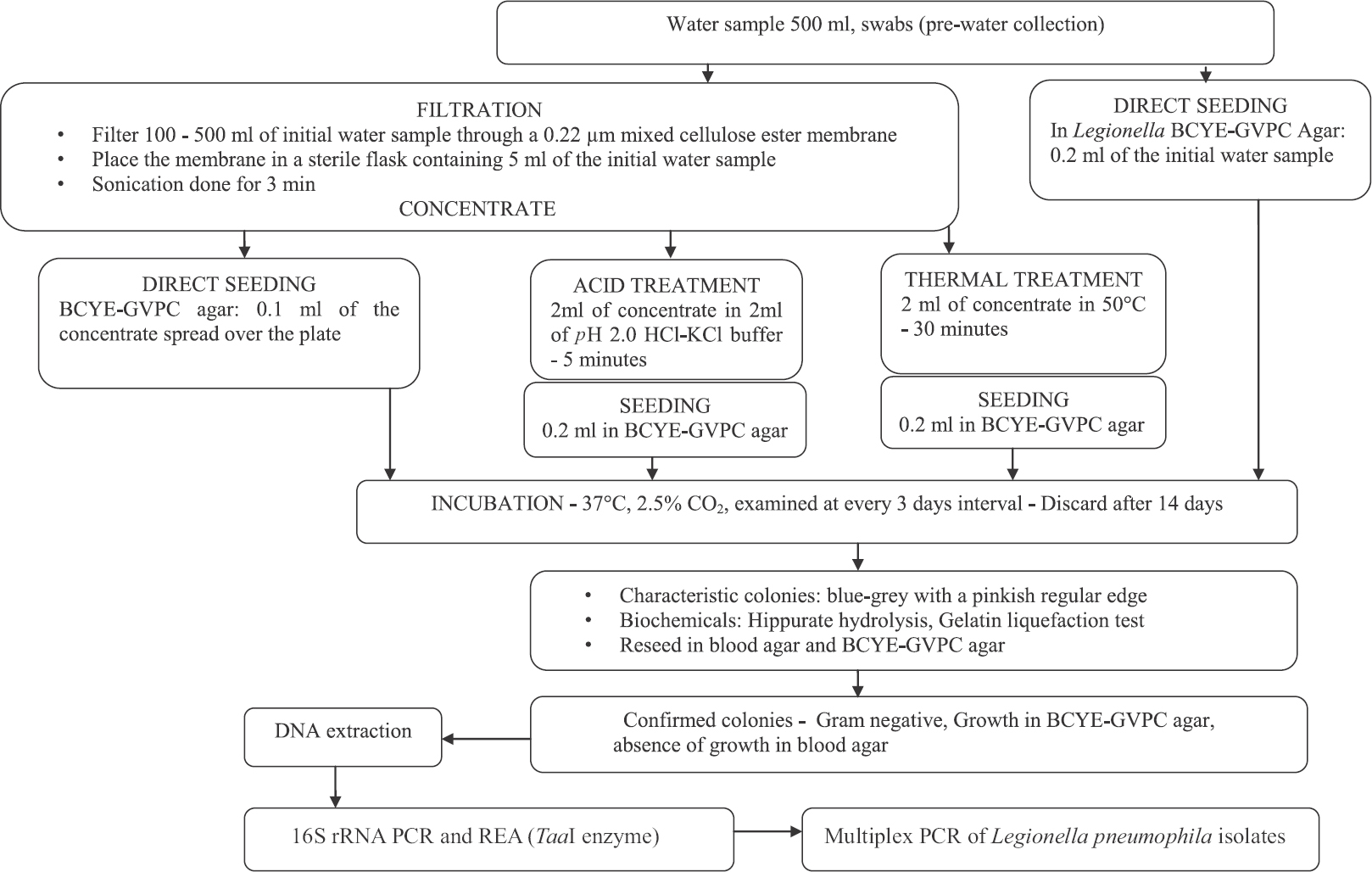
- Flowchart showing processing of water samples. BCYE-GVPC, buffered charcoal-yeast extract with Glycine, Vancomycin, Polymyxin B, and Cycloheximide; PCR, polymerase chain reaction; REA, restriction enzyme analysis.
Multiplex PCR of environmental isolates: The multiplex PCR for five markers was applied as mentioned above to randomly chosen 45 hospital environmental isolates of L. pneumophila and one non-pneumophila Legionella species. Electrophoresis of the products was done in 1.5 per cent agarose gel stained with ethidium bromide, visualized under UV light and gel documentation system (Gel Doc EZ imager, Bio-Rad, USA). The presence or absence of the band was compared with the bands of standard strain ATCC 33153.
Results
Culture, PCR and REA confirmation: The culture of standard strains showed characteristic ground glass appearance. These were Gram-negative bacilli and no growth was seen in blood agar. The 16S rRNA gene PCR showed 226 bp product which was digested into 180 bp and 46 bp by TaaI restriction enzyme to confirm it as L. pneumophila (Fig. 2).
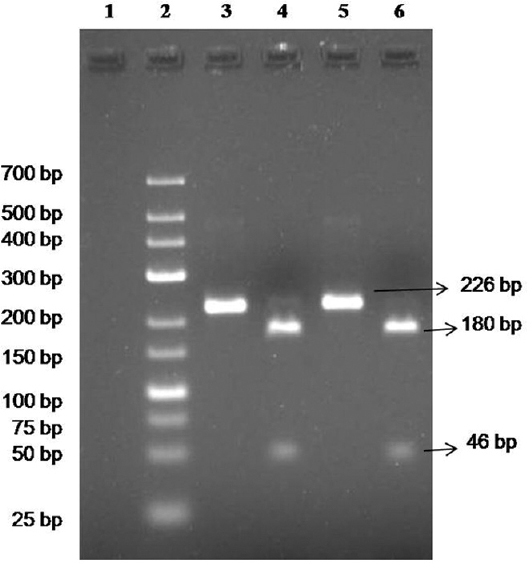
- 16S rRNA gene polymerase chain reaction (PCR) and restriction enzyme analysis (REA) of standard strains. Lane 1, negative control; lane 2, low range Marker (25-700 bp); lanes 3 & 4, PCR & REA of ATCC 33152 Legionella pneumophila, respectively; lanes 5 & 6, PCR & REA of ATCC 33153 L. pneumophila, respectively.
Multiplex PCR, sensitivity and specificity: The individual PCR for the markers showed expected product size, except for 11A2 marker. The PCR was found to be sensitive (Fig. 3A) to detect about 0.1 ng/μl of DNA and specific (Fig. 3B) for L. pneumophila standard strains used in the study. Both the ATCC standard strains of L. pneumophila used in the study showed a product at 200 bp region for marker 11A2. The ATCC strain 33153 showed the presence of all the five markers, whereas the marker 24B1 was not amplified in ATCC 33152. Sequencing of the gel-purified PCR products of ATCC strain 33153 showed 98-100 per cent identity with the GenBank sequences (https://www.ncbi.nlm.nih.gov/nucleotide) for the markers 19H4, 10B12, 24B1 and 8D6. The sequencing of marker 11A2 (pLPP) (~200 bp in our study) showed no significant similarity with L. pneumophila marker 11A2 available in the database (Gen bank Acc. no. HM584934). For marker 11A2, based on BLAST search (https://blast.ncbi.nlm.nih.gov/Blast.cgi) with the available database, it was found that 95-98 per cent identity was seen with other L. pneumophila strains such as strain thunder bay, ATCC 43290, 2300/99 Alcoy, strain Corby, strain Philadelphia and strain Paris annotating to HelA protein, cobalt/zinc/cadmium efflux RND transporter and HME protein importer family proteins.
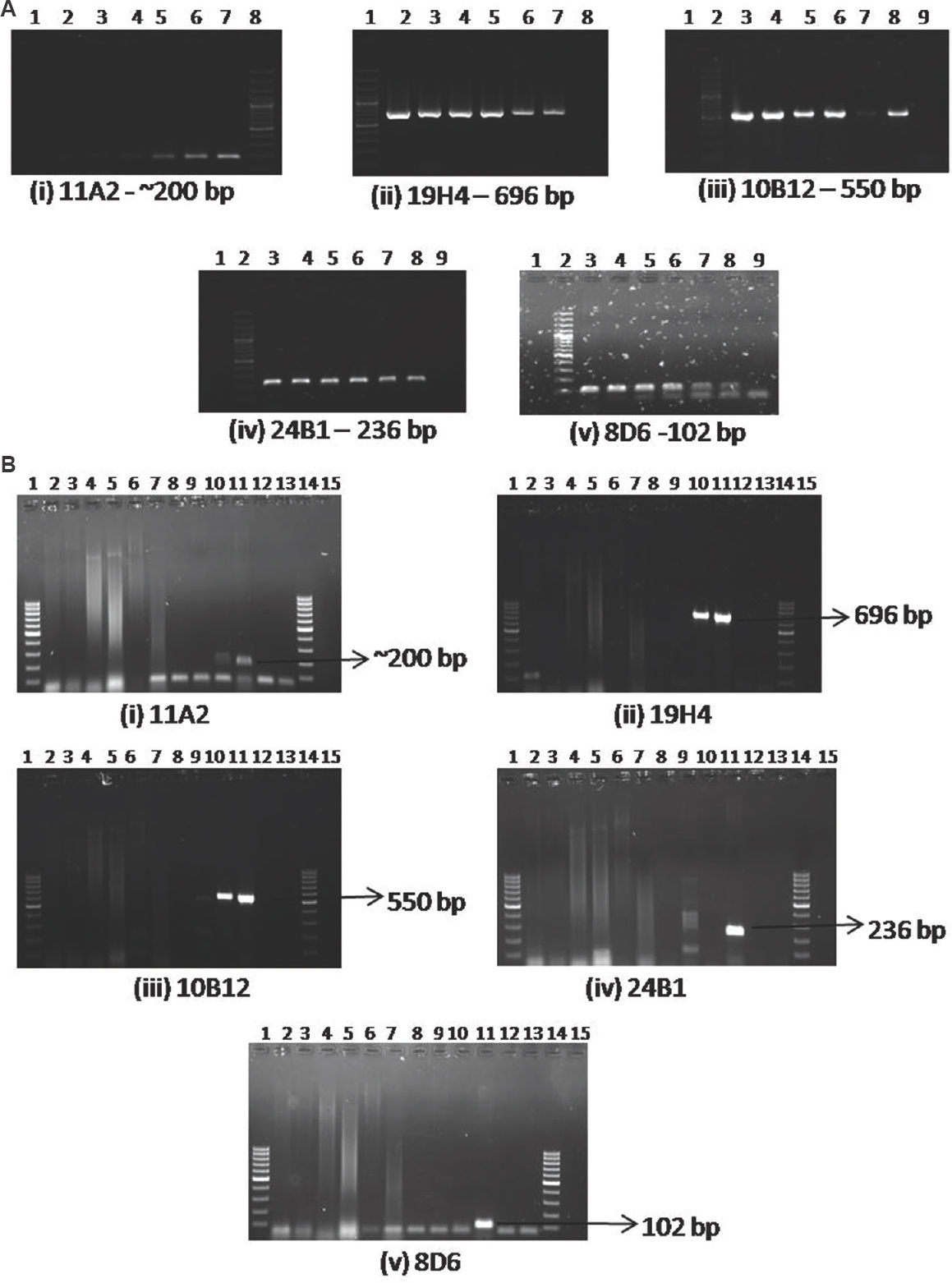
- (A) Sensitivity of PCR for individual markers: (i) 11A2 - Lane 1, negative control; lane 2, 1:1000 dilution (0.17 ng/μl); lane 3, 1:500 dilution; lane 4, 1:100 dilution; lane 5, 1:50 dilution; lane 6, 1:10 dilution; lane 7, ATCC 33153 DNA (177.4 ng/μl); lane 8, 100 bp ladder. (ii) 19H4 - Lane 1, 100 bp ladder; lane 2, ATCC 33153 DNA (177.4 ng/μl); lane 3, 1:10 dilution; lane 4, 1: 50 dilution; lane 5, 1:100 dilution; lane 6, 1:500 dilution, lane 7, 1:1000 dilution (0.17 ng/μl); lane 8, negative control. (iii) 10B12 - Lane 1, Blank; lane 2, 100 bp ladder; lane 3, ATCC 33153 DNA (177.4 ng/μl), lane 4, 1:10 dilution; lane 5, 1: 50 dilution; lane 6, 1:100 dilution; lane 7, 1:1000 dilution (0.17 ng/μl), lane 8, 1:500 dilution; lane 9, negative control. (iv) 24B1 - Lane 1, Blank, lane 2, 100 bp ladder; lane 3, ATCC 33153 DNA (177.4 ng/μl), lane 4, 1:10 dilution; lane 5, 1: 50 dilution; lane 6, 1:100 dilution; lane 7, 1:500 dilution; lane 8, 1:1000 dilution (0.17 ng/μl); lane 9, negative control. (v) 8D6 - Lane 1, Blank; lane 2, 100 bp ladder; lane 3, ATCC 33153 DNA (177.4 ng/μl); lane 4, 1:10 dilution; lane 5, 1: 50 dilution; lane 6, 1:100 dilution; lane 7, 1:500 dilution; lane 8, 1:1000 dilution (0.17 ng/μl); lane 9, negative control. (B) Specificity of PCR for individual markers: (i) 11A2, (ii) 19H4, (iii) 10B12, (iv) 24B1, (v) 8D6 - Lane 1, 100 bp ladder; lane 2, Escherichia coli DNA; lane 3, Klebsiella pneumoniae DNA; lane 4, Proteus mirabilis DNA; lane 5, Pseudomonas sp. DNA; lane 6, Haemophilus influenzae DNA; lane 7, Neisseria meningitidis DNA; lane 8, Staphylococcus aureus DNA; lane 9, Mycobacterium tuberculosis DNA; lane 10, Legionella pneumophila ATCC 33152 DNA; lane 11, L. pneumophila ATCC 33153 DNA; lane 12, negative control; lane 13, Blank; lane 14, 100 bp ladder.
Culture, PCR-REA and multiplex PCR of environmental isolates: In a span of 22 months, the bacterium was isolated on 30 instances (21 instances by water samples only and nine instances by swabs samples). On 29 instances, L. pneumophila was isolated, and on one instance, non-L. pneumophila species was isolated as confirmed by PCR and REA. Isolation of the bacterium was seen in samples with temperature as low as 13°C to as high as 47°C. The bacterium was isolated in 21.5 and 6.3 per cent of non-potable and potable drinking water, respectively.
Of the 45 hospital environmental isolates of L. pneumophila, 11 isolates had four markers, 12 isolates had three markers, 10 isolates had two markers, nine isolates had one marker and three isolates had none of the markers. In our study, none of the isolates showed all the five markers in the multiplex PCR. All the isolates had the expected product size as compared to the bands in the standard strain ATCC 33153. However, of the 25 isolates, in the context of 11A2 marker, 19 showed the expected product size of 355 bp (Table II) in contrast to the standard strain which showed a band in 200 bp region (Fig. 4). Fig. 5A and B shows the representative picture of multiplex PCR in isolate numbers 27-45. The DNA of the one non-pneumophila Legionella species isolated from environment tested by the multiplex PCR showed one band at 355 bp which is the product size of 11A2 marker.
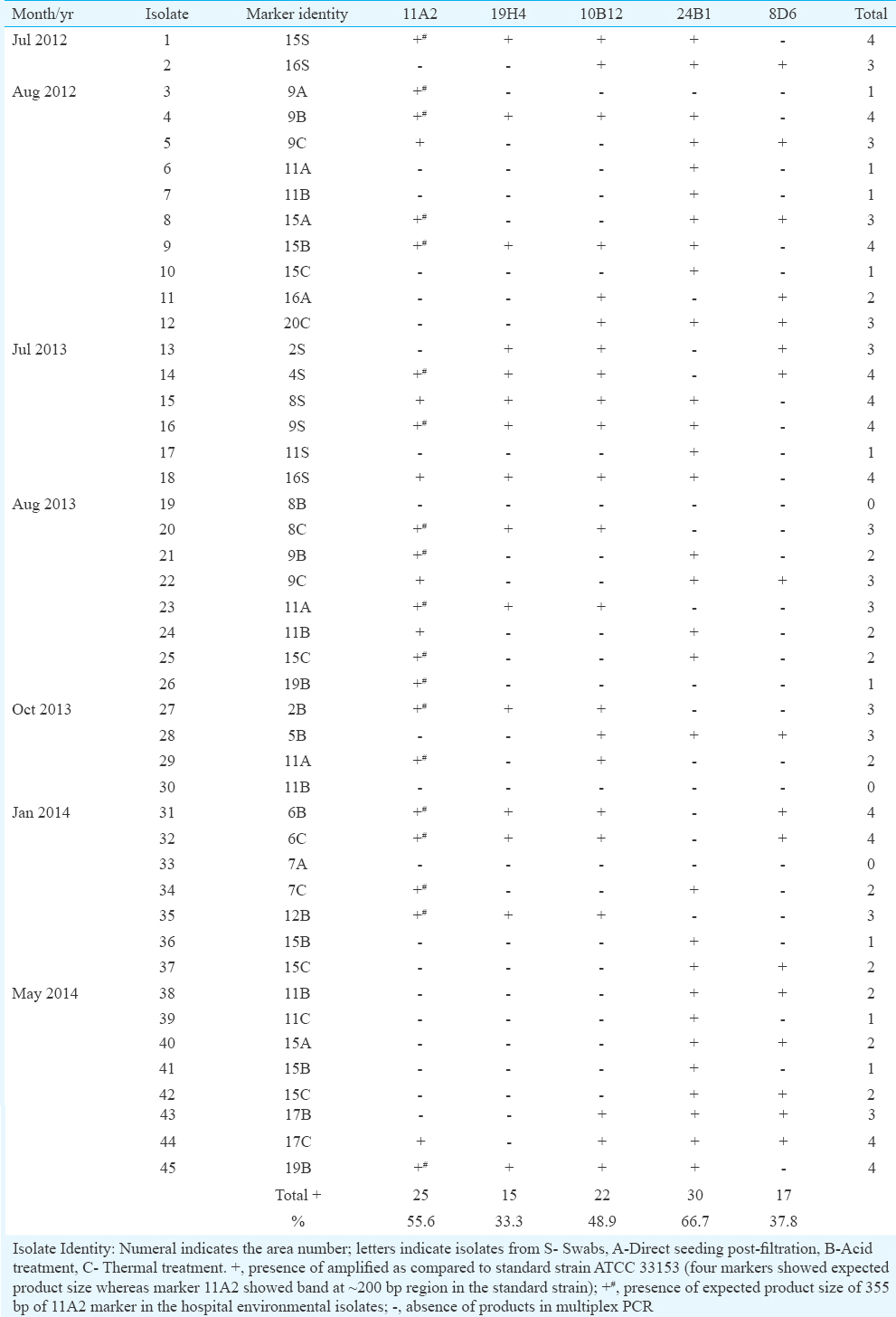
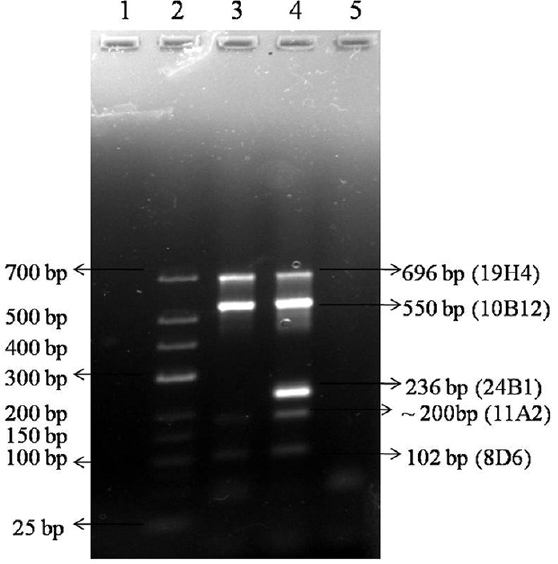
- Multiplex PCR of five markers in Legionella pneumophila standard strains ATCC 33152 and ATCC 33153 - Lane 1, Blank; lane 2, low range marker 25-700 bp size; lane 3, Legionella pneumophila ATCC 33152 DNA; lane 4, L. pneumophila ATCC 33153 DNA; lane 5, negative control.
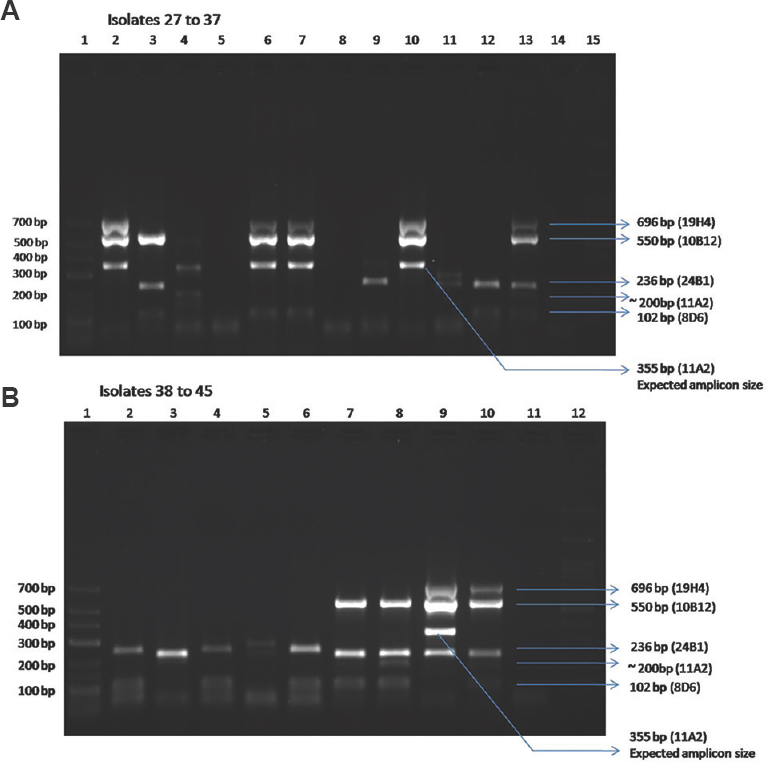
- Multiplex PCR of environmental Legionella pneumophila isolates (isolates 27 to 45 represented here). (A) Isolate 27 to 37 - Lane 1: low range Marker 25-700 bp, lane 2: isolate 27 (2B); lane 3, isolate 28 (5B); lane 4, isolate 29 (11A2); lane 5, isolate 30 (11B); lane 6, isolate 31 (6B); lane 7, isolate 32 (6C); lane 8, isolate 33 (7A); lane 9, isolate 34 (7C); lane 10, isolate 35 (12B); lane 11, isolate 36 (15B); lane 12, isolate 37 (15C); lane 13, positive control (ATCC 33153 Legionella pneumophila); lane 14, negative control; lane 15, 100 bp ladder. (B) Isolate 38 to 45 - Lane 1, low range Marker 25-700 bp; lane 2, isolate 38 (11B); lane 3, isolate 39 (11C), lane 4, isolate 40 (15A); lane 5, isolate 41 (15B); lane 6, isolate 42 (15C); lane 7, isolate 43 (17B); lane 8, isolate 44 (17C); lane 9, isolate 45 (19B); lane 10, positive control (ATCC 33153 L. pneumophila); lane 11, negative control; lane 12, 100 bp ladder.
Discussion
For epidemiological purposes, sequence-based typing scheme of Legionella isolates involving seven genes is followed worldwide12. Identification of genomic markers of pathogenic potential on Legionella bacterium is new.
The marker 24B1 (66.7%) was the most common in our isolates, followed by 11A2 (55.6%), 10B12 (48.9%), 8D6 (37.8%) and 19H4 (33.3%). The standard Philadelphia strain did not show the marker 24 B1. This means it is not necessary that the entire five markers be present together to give the strain a pathogenic potential. It is to be noted that the markers 24B1 and 11A2 are genes annotated to conjugative coupling factor and a plasmid, respectively, which may have a potential role in the horizontal gene transfer of the bacterium13. It is intriguing to note from our findings that in nine isolates where only one marker was found, seven isolates had 24B1 and two isolates showed 11A2 alone.
All the markers showed expected product size with the standard strains used in the study except the marker 11A2. Of the 45 L. pneumophila isolates studied, 25 isolates showed the presence of 11A2 marker (19 isolates showed an expected product size of 355 bp and six isolates showed a band in 200 bp region similar to the standard strain). This particular marker 11A2 is annotated to the plasmid of pLPP (Paris genome). The presence of an expected product for this plasmid in our isolates showed that this plasmid might not be specific to the Paris strain alone. Moreover, the presence of 11A2 marker in one non- L. pneumophila species isolated from our hospital showed that this plasmid was not specific for L. pneumophila but could be diversely present with other Legionella species also. Detailed studies for the presence of this plasmid as well as other markers in various Legionella species collected worldwide could give a better understanding of their ecological prevalence.
It is not necessary that the presence of these five markers in our isolates has proven their pathogenic potential because the ecological entities of continents are different. It is to be noted that the markers studied here may be applicable only to the European continent, especially in the Netherlands, for prediction, since genetic differences of the bacterium in different geographical areas have been documented14. Moreover, detailed mixed genome microarray studies have predicted different sets of markers in Dutch and French strains of L. pneumophila1516. A repository of gene markers identified for specific geographical areas may help in epidemiological studies.
It is generally accepted that when 30 per cent or more of the water samples show Legionella growth, the system can be considered as contaminated17. In our study, the presence of L. pneumophila was observed in 15.4 per cent of the hospital water distribution systems. Apart from the distal outlets, the bacterium was isolated on more than three instances from two AC towers installed in the hospital. The cooling towers were cleaned and freshwater was infused and flushed out twice without chemical dosing. Water samples collected from these sites on April 2015 after cleaning turned out to be negative for the bacterium when tested by culture. However, in May 2015, cultures from the sites turned positive again showing that routine cleaning and monitoring is necessary to control the bacterium at least for tertiary care centres.
The first report of Legionella isolation in India showed culture positivity in nine per cent of clinical samples and 76 per cent of environmental samples18. Our earlier studies showed a seroprevalence of about 15 per cent (IgM antibodies) and antigenuria of about 17 per cent in community-acquired pneumonia cases1920. Only one fatal case associated with community-acquired L. pneumophila infection in a known immunocompromised individual has been reported21. A study group from south India has shown a culture positivity rate of about two per cent in clinical sample and about 33 per cent in environmental samples2223.
Our study had certain limitations. Only the bacteria which were recovered by culture were assessed for the presence of markers. However, viable but non-culturable forms24 of this bacterium were not looked for and the specificity of the PCR with respect to other Legionella species was not checked. Moreover, it is essential to collect isolates from the patients and compare it with the environmental strains to get a clear picture of the infection-causing strains in India.
In conclusion, this study supports the idea that the distribution of genes of pathogenic potential in L. pneumophila may be diverse worldwide and rapid tests as the multiplex PCR reported here can help in epidemiological studies with respect to hospital infection control.
Acknowledgment
Authors thank Drs Arti Kapil, Benu Dhawan, Urvashi B. Singh of department of Microbiology, AIIMS, New Delhi, for their valuable suggestions and inputs. We thank Dr Urvashi B. Singh, department of Microbiology, AIIMS for providing DNA of M. tuberculosis. This study was funded by an intramural research grant from AIIMS, New Delhi (no. A-204).
Conflicts of Interest: None.
References
- Current and emerging Legionella diagnostics for laboratory and outbreak investigations. Clin Microbiol Rev. 2015;28:95-133.
- [Google Scholar]
- Distribution of Legionella species and serogroups isolated by culture in patients with sporadic community-acquired legionellosis: An international collaborative survey. J Infect Dis. 2002;186:127-8.
- [Google Scholar]
- Population structure and minimum core genome typing of Legionella pneumophila. Nat Sci Rep. 2016;6:21356.
- [Google Scholar]
- Comparison of clinical and environmental isolates of Legionella pneumophila obtained in the UK over 19 years. Clin Microbiol Infect. 2007;13:78-85.
- [Google Scholar]
- Hospital-acquired legionellosis: Solutions for a preventable infection. Lancet Infect Dis. 2002;2:368-73.
- [Google Scholar]
- Comparative genome analysis of a large Dutch Legionella pneumophila strain collection identifies five markers highly correlated with clinical strains. BMC Genomics. 2010;11:433.
- [Google Scholar]
- Two-step scheme for rapid identification and differentiation of Legionella pneumophila and non-Legionella pneumophila species. J Clin Microbiol. 2010;48:433-9.
- [Google Scholar]
- MPprimer: A program for reliable multiplex PCR primer design. BMC Bioinformatics. 2010;11:143.
- [Google Scholar]
- Centers for Disease Control and Prevention. Legionella (Legionnaires’ Disease and Pontiac Fever) Available from: https://www.cdc.gov/legionella/health-depts/inv-toolscluster/lab-inv-tools/procedures-manual.html
- [Google Scholar]
- Consensus sequence-based scheme for epidemiological typing of clinical and environmental isolates of Legionella pneumophila. J Clin Microbiol. 2005;43:2047-52.
- [Google Scholar]
- Extensive recombination events and horizontal gene transfer shaped the Legionella pneumophila genomes. BMC Genomics. 2011;12:536.
- [Google Scholar]
- Pan-European study on culture-proven Legionnaires’ disease: Distribution of Legionella pneumophila serogroups and monoclonal subgroups. Eur J Clin Microbiol Infect Dis. 2002;21:710-6.
- [Google Scholar]
- Genome analysis of Legionella pneumophila strains using a mixed-genome microarray. PLoS One. 2012;7:e47437.
- [Google Scholar]
- Prediction of the origin of French Legionella pneumophila strains using a mixed-genome microarray. BMC Genomics. 2013;14:435.
- [Google Scholar]
- Prevention of legionnaires’ disease in hospitals. Tidsskr Nor Laegeforen. 2011;131:1554-7.
- [Google Scholar]
- Isolation of Legionella pneumophila from patients of respiratory tract disease & environmental samples. Indian J Med Res. 1991;93:364-5.
- [Google Scholar]
- The incidence of Legionella pneumophila: A prospective study in a tertiary care hospital in India. Trop Doct. 2000;30:197-200.
- [Google Scholar]
- Sero diagnosis of Legionella infection in community acquired pneumonia. Indian J Med Res. 2010;131:92-6.
- [Google Scholar]
- Legionella pneumophila infection associated with renal failure causing fatality in a known case of sarcoidosis. Indian J Med Microbiol. 2014;32:324-7.
- [Google Scholar]
- Isolation of Legionella pneumophila from clinical & environmental sources in a tertiary care hospital. Indian J Med Res. 2010;131:761-4.
- [Google Scholar]
- Clinical prevalence of Legionella, associated risk and clinical features. Int J Biol Med Res. 2014;5:4582-5.
- [Google Scholar]
- Viability PCR, a culture-independent method for rapid and selective quantification of viable Legionella pneumophila cells in environmental water samples. Appl Environ Microbiol. 2009;75:3502-12.
- [Google Scholar]






