Translate this page into:
Multiple locus sequence typing of Salmonella Typhi, isolated in north India - a preliminary study
Reprint requests: Dr Arti Kapil, Professor, Department of Microbiology, All India Institute of Medical Sciences, Ansari Nagar, New Delhi 110 029, India e-mail: akapilmicro@gmail.com
-
Received: ,
This is an open-access article distributed under the terms of the Creative Commons Attribution-Noncommercial-Share Alike 3.0 Unported, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
This article was originally published by Medknow Publications & Media Pvt Ltd and was migrated to Scientific Scholar after the change of Publisher.
Abstract
Background & objectives:
In India enteric fever is a major public health problem and Salmonella Typhi is the most common aetiologic agent. Any control strategy for such infections depends to a large extent on the understanding of the disease and relatedness of strains across the world. Multi locus sequence typing (MLST) is one such method of genotyping of bacteria based upon housekeeping genes of known function and chromosome position. MLST data of pathogens are important to determine the molecular evolution by a stable and reproducible method. This study was undertaken to determine the sequence types of representatives S. Typhi isolates obtained from enteric fever patients in a tertiary care centre in north India, over a period of 20 years (1990-2010).
Methods:
A total of 30 representative isolates of S. Typhi identified by biochemical and serological tests were subjected to multi locus sequence typing (MLST). Seven housekeeping genes of known function and chromosome position were used for the typing by MLST. Sequencing was carried out by using an automated DNA sequencer and results were analyzed to generate phylogenetic tree.
Results:
MLST pattern grouped S. Typhi into two sequence types- ST1 and ST2. ST1 was predominantly present followed by ST2.
Interpretation & conclusions:
By MLST the presence of both sequence types, ST1 and ST2, was found in S. Typhi isolates in our region. Predominately ST1 was present followed by ST2. These preliminary results corroborate the global distribution of both sequence types of S. Typhi and also emphasize for the continuous screening of S. Typhi.
Keywords
Multi locus sequence typing
Salmonella Typhi
sequence type
typhoid fever
Despite the availability of a good vaccine and effective antibiotics, enteric fever remains to be an important public health problem and a major therapeutic challenge as antimicrobial resistance in Salmonella enterica serovar Typhi (S. Typhi) has led to limited antimicrobial choices123. Globally, 22 million typhoid cases occur annually and result in 6, 00,000 deaths approximately accounting highest concentration in Asia, especially in the Indian subcontinent4. To study the dynamics of spread of a disease and to control its antimicrobial resistance; it is necessary to type the strains by a method, which is discriminatory yet conservative and comparable across the laboratories so as to understand the evolutionary relationship within the serotypes. This is relevant in the present day scenario because infectious diseases are no longer confined to a particular geographical area.
Many studies have reported the genetic diversity within S. Typhi population using pulsed-field gel electrophoresis (PFGE) during outbreaks5. However, in endemic areas multiple PFGE genotypes circulate which may not be informative for the evolutionary relationships of the isolates across the globe5. It has been reported earlier that S. Typhi genome can differ up to 20 per cent in different isolates6. Also serovar S. Typhi has been shown to be variable due to homologous recombination between ribosomal RNA genes7.
Kidgell et al8 have described a set of seven housekeeping genes for the analysis of sequence diversity in Salmonella enterica and reported that multiple locus sequence typing (MLST) can be used for the population structure study and for detecting genetically related clones in S. Typhi. This information is lacking from Indian subcontinent where enteric fever is a major public health problem. MLST database is being generated all over the world for most of the pathogenic bacteria91011 but presently very limited information is available on S. Typhi MLST scheme especially for the Indian isolates. Till date, only three isolates from Indian subcontinent have been studied8. We, therefore, carried out MLST using the 7 housekeeping genes to determine the sequence types of 30 S. Typhi representative isolates obtained from patients of enteric fever admitted in a tertiary care hospital in New Delhi, north India, over a period of 20 years.
Material & Methods
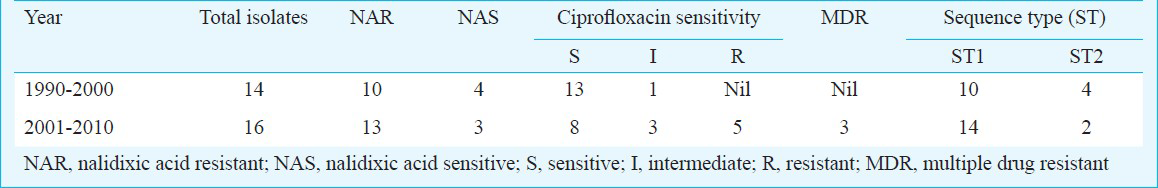
Bacterial isolates: A total of 30 representative isolates of S. Typhi obtained from the patients suffering from enteric fever admitted to the All India Institute of Medical Sciences (AIIMS), New Delhi, India over a period of 20 years (1990-2010), were included in the study (Table I). All isolates were identified by standard biochemical tests2 and serotyped by using specific antisera (dH and O9) (Murex Diagnostics Ltd, UK). The isolates were stored in glycerol at -70 oC and antimicrobial susceptibility was performed after revival and confirmation of the isolates by biochemical and serological tests.
Antimicrobial susceptibility was determined by disk diffusion method according to the Clinical Laboratory Standards Institute (CLSI) guidelines, 201012. Isolates resistant to ampicillin, chloramphenicol and co-trimoxazole were defined as multi drug resistant (MDR). MIC (minimum inhibitory concentration) for ciprofloxacin and ceftriaxone was determined by E-Test according to manufacturers’ instructions (AB Biodisk, Sweden).
The study was conducted in the Department of Microbiology and the study protocol was approved by Institute ethical committee.
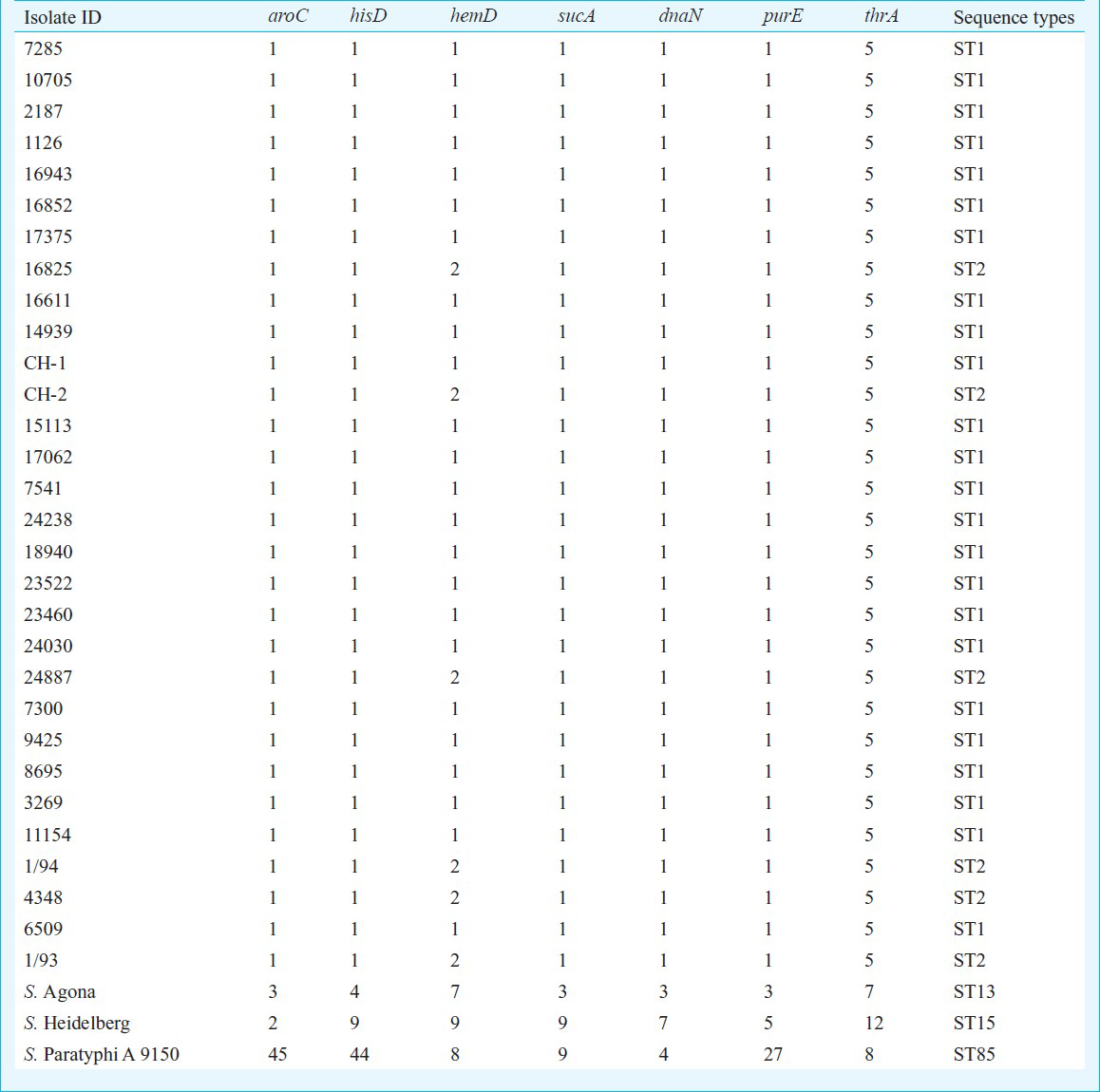
Multi locus sequence typing: Following seven housekeeping genes of known function and chromosome position, thrA (aspartokinase+homogenize dehydrogenase), purE (phosphoribosylaminoimidazole carboxylase), sucA (alpha ketoglutarate dehydrogenase), hisD (histidinol dehydrogenase), aroC (chorismate synthase), hemD (uroporphyrinogen III cosynthase), and dnaN (DNA polymerase III beta subunit) were used for MLST8. Primer and PCR conditions were available on MLST database (http://mlst.ucc.ie/mlst/dbs/Senterica/documents/primersEnterica_html). Briefly, four to six colonies from the 18-24 h old culture grown were suspended in 100 μl of sterile redistilled water and vortexed to form a suspension. The suspension was boiled at 100 oC for 10 min and an equal volume of chloroform: isoamyl alcohol (24:1) solution was added to it. The suspension was centrifuged at 5300 × g for 10 min. The supernatant containing bacterial DNA was aspirated and used as template for PCR. Template DNA prepared from bacterial isolates was amplified by PCR with the use of oligonucleotide sequence for seven housekeeping genes (available in MLST database). All the oligonucleotide sequences for PCR as well as for sequencing are available in the database. The PCR was performed in a final reaction volume of 50 μl containing 5 μl of 10X polymerase buffer, 2.65U of Taq DNA polymerase. The PCR amplified DNA of segment were electrophoresed in 1.5 per cent (w/v) agarose gel (Life Technologies, GibcoBRL, Scotland) prepared in 0.5 x Tris- borate ethylenediamine tetra acetic acid buffer (Sigma-Aldrich Pvt Ltd., India), along with the DNA molecular weight marker (100 bp DNA ladder) (Banglore Genie Pvt Ltd., India). The PCR product was observed after staining agarose gel with ethidium bromide (0.5 μg/ml) by using a ‘ChemiImager Ready’ gel documentation system (Alpha Innotech Corporation, California, USA) and the gels were photographed using Gel Doc™ (Bio-Rad, Hercules, Calif, USA).
Linear amplification process (Cycle sequencing): Sequencing was carried out by the dideoxynucleotide chain termination method2, using an automated DNA sequencer ABI PRISM ® 310 Genetic Analyzer (Applied Biosystems, USA) using AmpliTaq Gold DNA polymerase (Applied Biosystems, USA) which is a modified form of AmpliTaq DNA polymerase.
The PCR2 profile for cycle sequencing was set up as initially rapid thermal ramp to 96°C for 10 sec followed by rapid thermal ramp to 50°C for five sec and rapid thermal ramp to 60°C for four min. This was followed by rapid thermal ramp to 4°C until the product was refrigerated. Big Dye® Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems, USA) was used for cycle sequencing on ‘Gene Amp PCR system 2400’ thermal cycler (Applied Biosystems, Foster City, California, USA).
Briefly, each 0.25 x reaction was prepared by adding Big Dye Terminator Ready Reaction mix (2.0 μl), 5x sequencing buffer (3.0 μl), PCR product DNA (template 60 to 80 ng), primer 1 μl (3.2 pmoles) and deionized water to make up the volume to 20 μl per reaction. The contents were mixed well by flicking the tube and spun briefly. The tubes were placed in the thermal cycler and volume set to 20 μl.
Sequence analysis: For phylogenetic relationships among S. Typhi isolates, forward and reverse DNA sequences were assembled, trimmed, edited and analyzed for each gene fragment. The standard sequences for alignment were taken from MLST database. Multiple alignments were done using Genedoc Multiple sequence alignment editor and shading utility version 2.6.00213 and Clustal X 1.8114. The merged sequences were used to generate phylogenetic tree using the unweighted pair group method with arithmetic averages (UPGMA) by using MEGA4 v4.115.
Results
Thirty S. Typhi isolates were analysed by MLST. To characterize other phenotypic characters of these isolates, antibiotic sensitivity pattern was determined which showed three isolates as MDR (resistant to ampicillin, co-trimoxazole and chloramphenicol) and 23 were nalidixic acid resistant (NAR). All were sensitive to ceftriaxone (MIC range 0.016-0.50 μg/ml), five were ciprofloxacin resistant and four ciprofloxacin intermediate while 21 isolates were ciprofloxacin sensitive (MIC range 0.016 to >32 μg/ml) (Table I). No association was found between MDR/ciprofloxacin resistant and and MLST types. ST (sequence type) was assigned based on the allelic profile. The seven housekeeping genes were concatenated for all isolates and an UPGMA tree was constructed. S. Agona, S. ParatyphiA ATCC 915016 and S. Heidelberg were taken as outgroup. Based on concatenated sequence of 6 housekeeping genes, all S. Typhi isolates were uniform. However, all isolates were mutated at one site in hemD gene resulting in non-synonymous changes in the gene product. All 30 S. Typhi isolates showed monophyletic lineage (Fig.) and clustered in to two sequence types - ST1 and ST2- in S. Typhi and at separate lineage for S. Agona, S. Paratyphi A and S. Heidelberg. MLST results showed that both the sequence types were circulating in India, predominately ST1 (Tables I and Tables II. MLST results have been submitted to MLST database (http://mlst.ucc.ie/mlst/dbs/Senterica/GetTableInfo_html).

- Genetic relationship as inferred using the UPGMA method. The optimal tree with the sum of branch length = 0.03227106 is shown. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1000 replicates) is shown next to the branches. The tree is drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree.
Discussion
Typhoid fever remains an important public health problem in India. Outbreaks of typhoid have been reported from Maharashtra17 Bangalore18 Chandigarh19 and Pondicherry20 in India. With increase in the travel and economic development, the disease is no longer limited to a geographical area. So to understand the global epidemiology, strain circulation in country and to devise control strategy, it is important to type the bacterial strains by a method, which is portable, comparable, discriminatory and reproducible over the years.
In recent years, phenotypic and genotypic typing methods have been used for classification or sub typing of S. Typhi. Phenotypic typing includes phage typing19 which is reproducible but lacks discrimination. Antibiogram is reproducible, amenable to automation but lacks discriminatory power. Genotyping has been done in S. Typhi by two different ways PFGE and MLST. In PFGE highly variable region within the bacterial population are identified to detect micro variation for outbreak investigation21. However PFGE may not be useful in typing sporadic cases. None of these methods provides appropriate information to infer phylogenetic relationships among Salmonella isolates and subtypes22. MLST detects the variation that accumulates very slowly in the population and likely to be selectively natural8. This is a sequence-based method, which provides an alternative approach for typing microbes on the basis of sequence diversity23. This method is now considered as a gold standard in the characterization of bacterial strains and can be used to ascertain the clonality across the geographical regions. In our study a non-synonymous change in the hemD gene present within 24 isolates was assigned to sequence type ST1. While 6 isolates possessed identical alleles at all seven loci were assigned to sequence type ST2 (Table II). Also, we included S. Typhi isolates collected over 20 years considering the fact that within a year, possibility of significant variation is small. In previous study8, ST2 was found from the Eurasia, South America and Africa as well as from India from 1918-1999 and 1981-2000, respectively. The presence of ST1 predominantly in India indicates that S. Typhi circulation might have changed from previous sequence type ST2 to ST1 over time; may be due to the increased international travel or undergoing neutral accumulation of sequence variants in housekeeping genes.
ST3 and ST8 were not found in the Indian isolates in the present study. All four sequence types (ST1, ST2, ST3 and ST8) were present in isolates from Africa8. ST3 (synonymous change in thrA gene) was assigned to the isolate SARB64 obtained from Senegal and ST8 (synonymous change in hisD gene) was assigned to the isolate tested from Zaire8. S. Agona, S. Heidelberg and S. Paratyphi 9150 belong to separate sequence types. More recent work has used single nucleotide polymorphism (SNP) analysis to further detail the epidemiology of infection, which is a high throughput method and has improved sensitivity24.
In conclusion, MLST was found to be a valuable tool in international and national surveillance and generation of reference data. Two sequence types ST1 and ST2 were found in our isolates as in Eurasia, South America and Africa. Although the present study had limited number of isolates from one centre only but has generated pilot data. There is a need to generate this kind of information for a large number of representative strains from all parts of India.
Acknowledgment
This work was partially supported by the Indian Council of Medical Research, New Delhi.
References
- S. Typhi with transferable chloramphenicol resistance isolated in Chandigarh during 1983-87. Indian J Pathol Microbiol. 1994;37:179-83.
- [Google Scholar]
- Reduced susceptibility to ciprofloxacin and gyra gene mutation in North Indian strains of Salmonella enterica serotype Typhi and serotype Paratyphi A. Microb Drug Resist. 2004;10:146-53.
- [Google Scholar]
- Antibiogram pattern and seasonality of Salmonella serotypes in a North Indian tertiary care hospital. Epidemiol Infect. 2006;134:961-6.
- [Google Scholar]
- Epidemiologic analysis of sporadic Salmonella typhi isolates and those from outbreaks by pulsed-field gel electrophoresis. J Clin Microbiol. 1994;32:1135-41.
- [Google Scholar]
- Genome size variation among recent human isolates of Salmonella typhi. Res Microbiol. 1997;148:229-35.
- [Google Scholar]
- Rearrangements in the genome of the bacterium Salmonella typhi. Proc Natl Acad Sci USA. 1995;92:1018-22.
- [Google Scholar]
- Salmonella Typhi, the causative agent of typhoid fever, is approximately 50,000 years old. Infect Genet Evol. 2002;2:39-45.
- [Google Scholar]
- Multilocus sequence typing system for Campylobacter jejuni. J Clin Microbiol. 2001;39:14-23.
- [Google Scholar]
- Multilocus sequence typing system for group B streptococcus. J Clin Microbiol. 2003;41:2530-6.
- [Google Scholar]
- Multilocus sequence typing for characterization of clinical and environmental Salmonella strains. J Clin Microbiol. 2002;40:1626-35.
- [Google Scholar]
- Performance Standards for Antimicrobial Susceptibility Testing. In: Supplement (June 2010 update) M100- S20, vol. 30 no. 15. Wayne, PA: CLSI; 2010.
- [Google Scholar]
- GeneDoc: analysis and visualization of genetic variation. 1997. Multiple sequence alignment editor & shading utility version 27.000 used offline. Available from: http://www.psc.edu/biomed/genedoc
- [Google Scholar]
- The CLUSTAL_X windows interface: Flexible strategies for multiple sequence alignment aided by qualityanalysis tools. Nucleic Acids Res. 1997;25:4876-82.
- [Google Scholar]
- MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596-9.
- [Google Scholar]
- Comparison of genome degradation in Paratyphi A and Typhi, human-restricted serovars of Salmonella enterica that cause typhoid. Nat Genet. 2004;36:1268-74.
- [Google Scholar]
- Epidemiological investigation of an outbreak of enteric fever in a village in Maharashtra. J Commun Dis. 1996;28:117-21.
- [Google Scholar]
- An outbreak of multidrug resistant typhoid fever in Bangalore. Indian J Pediatr. 1995;62:445-8.
- [Google Scholar]
- An outbreak of typhoid due to multidrug resistant Salmonella typhi in Pondicherry. Trans R Soc Trop Med Hyg. 1992;86:204-5.
- [Google Scholar]
- Antimicrobial resistance trends in blood culture positive Salmonella Typhi isolates from Pondicherry, India, 2005-2009. Clin Microbiol Infect. 2012;18:239-45.
- [Google Scholar]
- Multilocus sequence typing supports the hypothesis that cow- and human-associated Salmonella isolates represent distinct and overlapping populations. Appl Environ Microbiol. 2006;72(12):7575-85.
- [Google Scholar]
- Multilocus sequence typing: a portable approach to the identification of clones within populations of pathogenic microorganisms. Proc Natl Acad Sci USA. 1998;95:3140-5.
- [Google Scholar]
- Temporal fluctuation of multidrug resistant Salmonella Typhi haplotypes in the Mekong River Delta Region of Vietnam. PLoS Negl Trop Dis. 2011;5:1-10.
- [Google Scholar]






