Translate this page into:
Multiple drug resistant carbapenemases producing Acinetobacter baumannii isolates harbours multiple R-plasmids
The first two authors contributed equally
Reprint requests: Dr Prashanth K, Assistant Professor, Department of Biotechnology, School of Life Sciences Pondicherry University, R. Venkataraman Nagar, Kalapet, Puducherry 605 014, India e-mail: prashi2k@gmail.com
-
Received: ,
This is an open-access article distributed under the terms of the Creative Commons Attribution-Noncommercial-Share Alike 3.0 Unported, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
This article was originally published by Medknow Publications & Media Pvt Ltd and was migrated to Scientific Scholar after the change of Publisher.
Abstract
Background & objectives:
The nosocomial human pathogen Acinetobacter baumannii has high propensity to develop resistance to antimicrobials and to become multidrug resistant (MDR), consequently complicating the treatment. This study was carried out to investigate the presence of resistant plasmids (R-plasmids) among the clinical isolates of A. baumannii. In addition, the study was performed to check the presence of common β-lactamases encoding genes on these plasmids.
Methods:
A total of 55 clinical isolates of A. baumannii were included in the study and all were subjected to plasmid DNA isolation, followed by PCR to check the presence of resistance gene determinants such as blaOXA-23, blaOXA-51, blaOXA-58 and blaIMP-1 on these plasmids that encode for oxacillinase (OXA) and metallo-β-lactamase (MBL) type of carbapenemases. Plasmid curing experiments were carried out on selected isolates using ethidium bromide and acridine orange as curing agents and the antibiotic resistance profiles were evaluated before and after curing.
Results:
All the isolates were identified as A. baumannii by 16SrDNA amplification and sequencing. Plasmid DNA isolated from these isolates showed the occurrence of multiple plasmids with size ranging from 500bp to ≥ 25 kb. The percentage of blaOXA-51 and blaOXA-23 on plasmids were found to be 78 and 42 per cent, respectively and 20 isolates (36%) carried blaIMP-1 gene on plasmids. Significant difference was observed in the antibiograms of plasmid cured isolates when compared to their parental ones. The clinical isolates became susceptible to more than two antibiotic classes after curing of plasmids indicating plasmid borne resistance.
Interpretation & conclusions:
Our study determined the plasmid mediated resistance mechanisms and occurrence of different resistance genes on various plasmids isolated from MDR A. baumannii. The present findings showed the evidence for antibiotic resistance mediated through multiple plasmids in A. baumannii clinical isolates. This indicates towards a need for preventive measures to avert the dissemination of plasmid resistance determinants in clinical environments.
Keywords
Acinetobacter baumannii
carbapenem resistance
R-plasmids
Acinetobacter baumannii is responsible for various nosocomial infections that mainly affect critically ill patients in intensive care units (ICUs) and its most striking manifestation is endemic and epidemic occurrence as multiple drug resistant (MDR) strains in the hospitals across the globe12. Generally, carbapenems are the most active agents against A. baumannii, but carbapenem-resistant A. baumannii (CRAB) isolates are becoming increasingly common where the predominant resistance mechanism involves the expression of β-lactamases1 (either metalloenzymes or class D enzymes). Regardless of recent progress in knowledge of antibiotic resistance machinery in A. baumannii, very little is known about the genetic factors driving the isolates towards multiple-drug resistance. Though many genetic means of resistance to multiple classes of antibiotics are known to exist in this organism, the major ones appear to be intrinsic or acquired via plasmids, integrons, and transposons1. However, the mechanisms of the dissemination of mobile elements bearing β-lactamase genes between Acinetobacter spp. are not fully understood. Many earlier investigations have given special emphasis for plasmid-mediated transferable antibiotic resistance3. Transfer of R-plasmids from A. baumannii to other nosocomial pathogens can create complications during the treatment. Moreover, recognition of plasmid transfer is crucial for control of outbreaks caused by MDR nosocomial pathogens. Lack of sufficient reports from India on the molecular basis of resistance in A. baumannii necessitated this study. We examined the presence of R-plasmids in the clinical isolates of A. baumannii and checked for the existence of different resistance genes on these plasmids.
Material & Methods
This study was done at Department of Biotechnology, School of Life Sciences, Pondicherry University, Puducherry.
Bacterial isolates: Overall, 55 consecutive A. baumannii isolates were obtained from Pondicherry Institute of Medical Sciences hospital, Puducherry, India, from various clinical specimens like endotracheal aspirates, cerebrospinal fluid, wound swabs, urine and blood culture specimens from patients mostly admitted in ICUs and acute medical care units during 2008-2009. The isolates were characterized to the species level using standard microbiological tests and simplified phenotypic tests as described elsewhere4 and their identification was reconfirmed using 16s rDNA amplification and sequencing5.
Plasmid DNA isolation: The plasmids from A. baumannii were isolated by following the standard procedure with slight modifications3. Briefly, a single colony of the bacteria was inoculated into 10 ml of Luria Bertani (LB) broth and incubated overnight at 37°C. The cells were harvested by centrifuging at 13600 g for 5 min and the pellet was resuspended in 700 μl of solution I [50 mM glucose, 25 mM Tris Cl (pH 8.0), 10 mM EDTA; autoclaved and stored at 4°C] and mixed thoroughly by vortexing. Subsequently, 1 ml of solution II (0.2N NaOH, 1% SDS; freshly prepared from the stocks of 1N NaOH and 10% SDS) was added and the preparations were stored in ice for 5 min, followed by addition of 500 μl of ice-cold solution III [5M potassium acetate (60 ml), 11.5 ml glacial acetic acid (3M K+; 5M CH3COO-) in 28.5 ml distilled water; stored at 4°C)]. This preparation was mixed and stored in ice for 3 to 5 min and then centrifuged at 12000 g for 5 min and the supernatant was transferred to a fresh tube. Subsequently, 6 μl of RNase (Sigma, USA) (10 mg/ml) was added and the solution was incubated at 37°C for 30 min. Equal volumes of saturated phenol:chloroform (1:1 v/v) solution was added to this preparation, mixed well and centrifuged at 12000 g for 10 min. The resulting supernatant was transferred carefully into a fresh tube. Subsequently, twice the volume of ice-cold isopropanol was added to these tubes and stored at 4°C for overnight. The contents were centrifuged at 12000 g for 5 min, the supernatant was discarded and the pellet was rinsed well with 70 per cent ethanol, air-dried and resuspended in 100 μl of TE buffer (10 mM Tris HCl (pH 8.0), 1 mM EDTA, (pH 8.0).
Transformation experiments: Conjugation experiment was carried out as described elsewhere7 using A. baumannii isolates as the donor and Escherichia coli strain HB101, which was susceptible to most of the antibiotics tested, as the recipient. Plasmid transformation was checked through plasmid isolation followed by electrophoresis. Any trans-conjugants obtained were tested for antibiotic resistance to check for any acquired resistance. i.e. looking for resistance in the recipient strain for a particular antibiotic, which has already recorded resistance in the donor strain.
Plasmid-curing experiments: Ten clinical isolates that showed resistance to more than eight antibiotics including carbapenems were selected for further analysis by plasmid curing experiments. Acridine orange and ethidium bromide (EtBr) (Hi-Media, India) were used as curing agents and their minimal inhibitory concentrations (MICs) were determined for the isolates and the highest / sub-inhibitory concentration that permitted slight growth was used for plasmid curing (Acridine orange – 640 to 2560 μg/ml; EtBr – 320 to 5120 μg/ml). Curing broth was prepared by diluting curing agents to sub-inhibitory concentration using LB broth to a final volume of 10 ml. The test cultures were inoculated into the above curing broth, incubated for 48 h and plated on LB agar. The cured cultures were subjected to antibiotic susceptibility testing by inoculating them into 5 ml Mueller-Hinton (MH)-broth and incubating until the turbidity reaches 0.5 McFarland standard. Subsequently, antimicrobial susceptibility was checked by disc diffusion method as per Clinical Laboratory Standard Institute (CLSI) guidelines8 for different antibiotics, which were previously used to test before curing. MIC values of meropenem were also checked for cured isolates. Phenotypic metallo-β-lactamase (MBL) detection was carried out using the imipenem EDTA double disc synergy (DDS) test9. Production of oxacillinases was analyzed according to the method described by Zarilli et al10, wherein the assays were performed in Mueller-Hinton agar (MHA) plates with and without 200 mM of NaCl.
PCR screening for carbapenemase genes: All the isolated plasmids were checked for the presence of different genes encoding carbapenemases through PCR. The presence of predominant MBL gene namely blaIMP1 and the most common carbapenem-hydrolyzing class D β-lactamases coded by oxacillinase (OXA) type genes such as blaOXA-23, blaOXA-51 and blaOXA-58 were investigated. Primer sequences used for blaIMP-1, blaOXA-23, blaOXA-51 and blaOXA-58 amplifications are given in Table I. The in-house strains bearing all these four resistant determinants, confirmed by amplification and DNA sequencing, were used as positive controls for PCR. The PCR composition and reaction conditions employed were according to our earlier investigation11. All the MBL and oxacillinase positive isolates by both the methods were repeatedly checked for reproducibility.

Results
Majority of A. baumannii isolates were obtained from the wound specimen (n=24; 44%) whereas 29 per cent of isolates were from endotracheal aspirates (n=16). The remaining isolates were from urine (n=6) of patients with urinary tract infection (UTI), and from CSF (n=5) of patients suffering from secondary meningitis. Many wound infection patients have also shown blood culture positive for A. baumannii, when subsequent second sample was analyzed (n=13). However, at first only four isolates were directly from blood culture of patients suffering from blood stream infection (n=4) and were counted separately. All the 55 isolates were identified to be A. baumannii based on 16S rDNA sequencing followed by BLAST analysis.
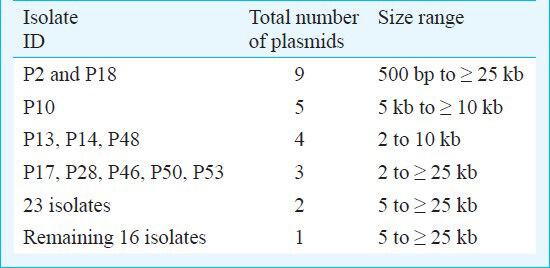
Presence of multiple R-plasmids: Plasmid profiles generated from the A. baumannii clinical isolates were analyzed and majority of the isolates had at least one plasmid. Of the 55 isolates tested, 50 (94.5%) contained plasmids. Isolates with identical plasmid profile were pooled in the same plasmid group and the variable number of plasmids (ranging from 1 to 9) with different molecular weights in each isolate enabled the identification of 10 plasmid profiles without the need for endonuclease digestion. The sizes of the plasmids were in the range of 2 to >25 kb (Fig. 1). Genotyping data of all the A. baumannii isolates that was previously performed and reported had generated six random amplified polymorphic DNA (RAPD) clusters (designated I-VI) revealing highly divergent distinct clones12. Four main groups of isolates comprised a total of 11 isolates showed the presence of three or more plasmids ranging from 500 bp to ≥ 25 kb with one common high molecular weight plasmid of size ≈ 12 or ≥ 25 kb (Table II). One major cluster of 23 isolates contained plasmids more than 5 kb except one isolate i.e. P43 which also had a 2 kb plasmid. The remaining group of 16 isolates contained only one plasmid with sizes ranging from 5 to ≥ 25 kb. Isolates such as P2 and P18 had highest number of plasmids. Multiple plasmids were common in majority of the isolates and only 16 isolates had single plasmid of high molecular-weight (>10 kb). Transformation experiments performed for native plasmid transformation showed that E. coli HB101 failed to acquire resistance to any of the antibiotics for which donor A. baumannii was resistant.
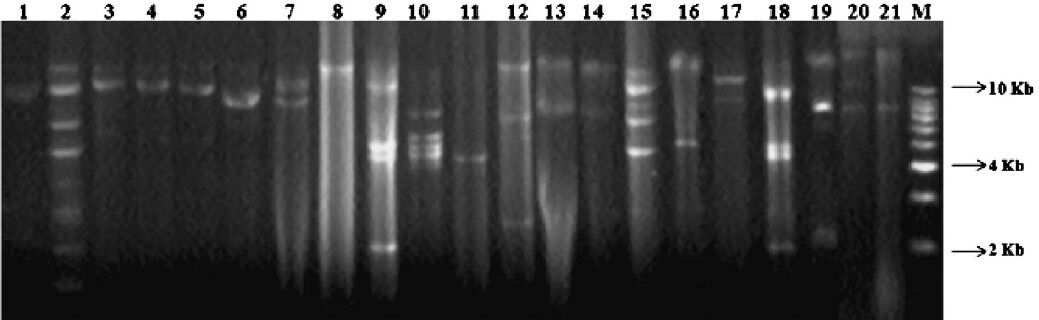
- Plasmid profiles of representative MDR A. baumannii isolates bearing single and multiple numbers of plasmids. (a) Lanes 1, 3-7, 17 (strain IDs P1, P4, P5, P6, P9, P11, P35) depict strains containing two plasmids of sizes greater than ≥ 10Kb and lane 16 containing two plasmids, one is of 5Kb and another of ≥ 10Kb size; (b) Lanes 13, 14 & 21 depict strains (P20, P3 & P23) containing two plasmids, one ≥ 10Kb and another of 8Kb size; (c) Lane 2 depicts strain (P2) containing multiple plasmids (n=9); (d) Lane 8 depicts strain (P12) harbouring single plasmid with molecular weight greater than 10Kb; Lane 11 depict strain (P16) harbouring single plasmid with molecular weight of 5Kb; (e) Lanes 9, 10 and 18 represents strains (P13, P14, P48) harbouring four plasmids of size varying from 2Kb to 10Kb; (f) Lane 12, 19 and 20 (strain IDs P17, P28, P22) contains three plasmids of size varying from 2Kb to >10Kb; (g) Lane 15 shows strain (P10) containing five plasmids of size varying from 5Kb to >10Kb.
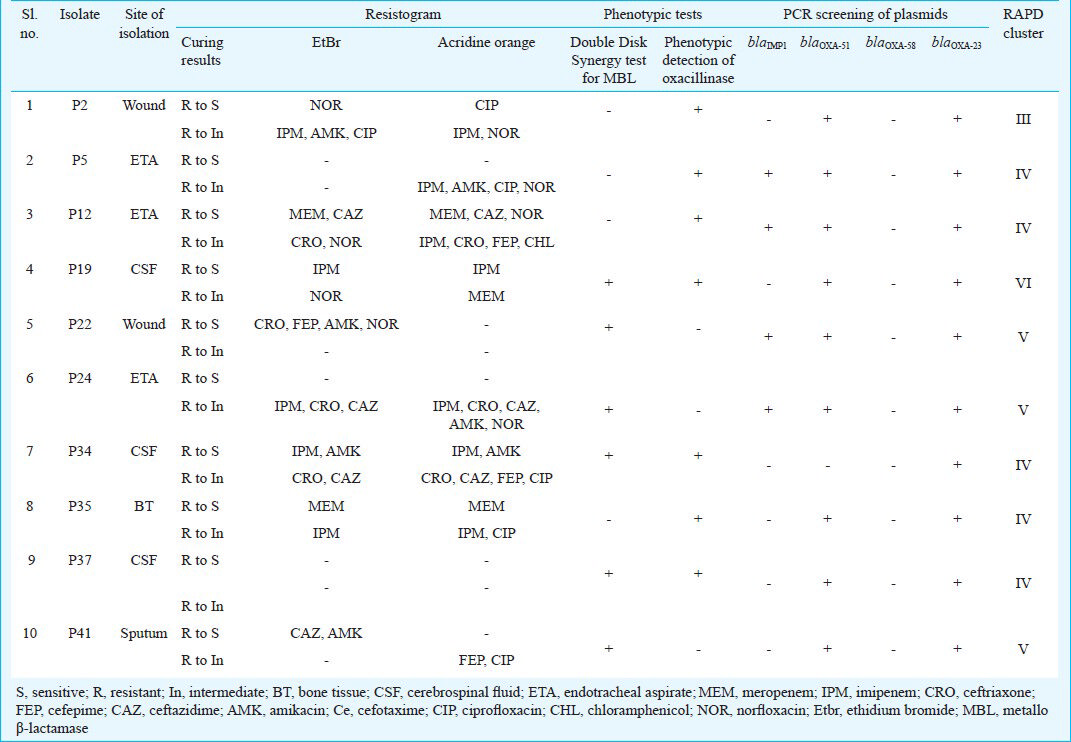
Resistance profiles after plasmid curing: Ten representative MDR clinical isolates were tested for ‘resistance’ properties by plasmid curing using known sub-inhibitory concentrations of curing agents and showed significant shift from resistance to susceptibility in nine of them when tested by disc-diffusion method. For instance, four isolates (P2, P22, P34 & P41) became susceptible to more than two antibiotics after EtBr curing. Conversely, P37 became resistant to imipenem and ceftriaxone after EtBr treatment, which was earlier susceptible. Most of the isolates became susceptible to more than two antibiotics when treated with acridine orange, and susceptibility to seven antibiotics (Table III). P12 showed four-fold reduction in its MIC for meropenem, P24 had a three-fold decrease, while P2 and P22 showed two-fold reductions in their MIC for meropenem after curing. Only P19 and P37 showed contrasting results wherein both became resistant to carbapenems and ceftriaxone after acridine orange curing.
Presence of OXA type and MBL genes: The results showed complete absence of blaOXA-58 both on the chromosome and plasmids. PCR results showed the presence of blaOXA-51 and blaOXA-23 on plasmids in around 39 (78%) and 21 (42%) isolates, respectively out of total 55 isolates tested. Twenty isolates (40%) had plasmids bearing blaIMP-1 gene (Fig. 2). PCR amplification and sequencing of IS element ISAba1 from representative isolates (n=6) confirmed the presence of ISAba1 upstream of OXA-carbapenemase genes, retaliating the association between OXA-genes and ISAba1. There was no association between the results of genotypic and phenotypic assays used to detect β-lactamases.
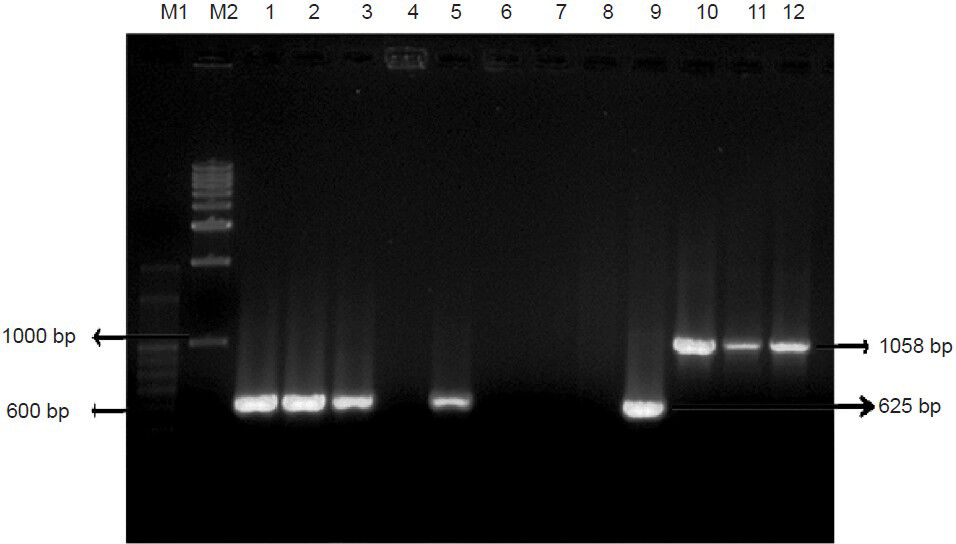
- PCR amplification of blaOXA-51 and blaOXA-23. Lane M1 - 100bp DNA ladder, Lane M2 - 1kb DNA ladder, Lane 1, 2, 3, 5 & 9 Positive amplification of blaOXA-51 (Isolate ID: P24, P25, P26, P28, P37); Lane 4, 6, 7 & 8 (P27, P29, P33, P36) Isolates showing no amplification of blaOXA-51; Lane 10-12 - Positive amplification of blaOXA-23 (P2, P19, P41).
Discussion
Acinetobacter genus appears to have high propensity to develop resistance in response to new antibiotics rapidly, unlike more “traditional” clinical bacteria2. The success of A. baumannii is attributed to its ability to respond quickly to new antibiotic challenges. Mobile genetic elements like plasmids and transposons play an important role in the biology of most prokaryotic organisms, and Acinetobacter species is no exception13. Plasmid profiling has been used to study the epidemiology of Acinetobacter in the past and >80 per cent of Acinetobacter isolates appear to carry multiple indigenous plasmids with variable molecular sizes1. Study conducted by Poirel et al14 conclusively inferred plasmids/integrons playing an important role in antibiotic resistance, endemic and epidemic behaviour of A. baumannii. Almost all our isolates harboured 1 to 2 high molecular weight plasmids (> 10 kb), which is in accordance with earlier observations. For instance, 63 kb plasmid pIP1841 of A. baumannii harbouring a gene, which confers resistance to aminoglycosides is self-transferable to other A. baumannii, A. haemolyticus and A. lwoffii but not to E.coli15.
All the isolates in this study were resistant to three or more antibiotics and plasmid curing experiments attributed resistance properties to one or more plasmids present in them. Two earlier Indian studies have documented A. baumannii bearing high molecular weight plasmids ranging from 5 to 66 kb716. One of them reported five different types of plasmids with a maximum of three plasmids in A. baumannii isolates with molecular size 1.5 to 40 kb and it also revealed that Acinetobacter spp. isolated from the tribal skin flora had low number of plasmids as compared to clinical and environmental isolates12. This study also documented that isolates harbouring low-molecular weight plasmids were susceptible to most of the antibiotics, which is contrary to our observations. The authors speculated that these smaller plasmids might be cryptic in nature or code for some other novel properties other than resistance16.
It is shown that Acinetobacter is capable of harbouring large plasmids in multiple numbers and these may be large due to the presence of many transferable integron cassettes in them, which are evidently responsible for resistance. Our PCR results showed the presence of blaIMP-1 gene only in larger plasmids. Earlier it has been revealed that the blaIMP-1 gene normally resides in a plasmid-borne class-1 integron cassette114. Numerous studies have noted integron cassettes on larger plasmids, which is likely to be true in our isolates also. The presence of a common large size plasmid (≈25-40kb) among all our isolates possibly indicates the presence of transferable integrons cassettes responsible for resistance.
Plasmid-mediated transfer of blaNDM-1 gene in Gram-negative bacteria was investigated in a study in which five blaNDM-1 positive plasmids of different incompatibility groups from clinical isolates belonging to Enterobacteriaceae were evaluated for conjugation properties and host specificity. Successful conjugative transfers were obtained between members of enterobacterial spp. when these were used as either donor or recipients. However, none of the five plasmids were transferable to A. baumannii17. Likewise, we failed to obtain transconjugants when E. coli HB101 was used as a recipient. Though many intrinsic Acinetobacter plasmids reported are non-self-transferable, the almost ubiquitous replicase gene appears to be strongly associated with a potential tra locus that could serve as a general system for plasmid mobilization. Consequently, horizontal gene transfer (HGT) of resistance genes among A. baumannii strains may not be ruled out18. Therefore, one can presume that successful resistance determinant transfer occurs between two Acinetobacter strains and transfer of plasmids directly from a member of enterobacteriacae to acinetobacter or vice versa is difficult or a rare event.
Earlier it was noted that EtBr curing led to disappearance of resistance to extended spectrum of β-lactams (ESBL) with the concurrent loss of plasmids from A. baumannii4 suggesting that ESBL determinants were plasmid-borne. Similarly, a significant shift was observed in our study from resistance to susceptibility as well as loss of plasmids after curing indicating that most of the clinical isolates had plasmids bearing multiple resistance determinants on them. Four of the 10 isolates became susceptible to more than two antibiotics after curing and isolates P2, P22 and P41 became susceptible to three antibiotic classes such as carbapenem, aminoglycoside and quinolone and/or cephalosporin.
Presence of blaOXA-51, blaOXA-23 and blaIMP-1 was predominantly observed on plasmids in our study and these resistant determinants can be easily transferred to other isolates of A. baumannii. Plasmid-mediated HGT of common blaOXA-24/40-carrying plasmids (30-kb repA_AB; 10-kb aci2) coding for imipenem resistance was recently demonstrated in A. baumannii and here the blaOXA-24/40 was also identified in the chromosome19. OXA-40 present on Pseudomonas aeruginosa plasmid showed 100 per cent homology to blaOXA-24/40 carried by A. baumannii plasmid. Further, A. radioresistens, a commensal isolated from the skin has been identified as the source for this OXA-4020. Epidemiologically unrelated CRAB isolates belonging to European clonal lineage-II, have been shown to carry an identical OXA-24-encoding plasmid pABVA0121. Interestingly, pABVA01 is flanked by conserved inverted repeats which are homologous to flanking regions of other Acinetobacter plasmids suggesting their mobilization is through a novel site-specific recombination mechanism.
It has been reported that CRAB bearing blaOXA-24 on plasmid expressing class-D carbapenemase releases it into outer membrane vesicles during in vitro growth that serves as a mechanism of HGT imparting the carbapenem resistance22. A plasmid-borne ISAba1-blaOXA-51-like gene along with its additional copy on their chromosomes has been identified in carbapenem resistant Acinetobacter genomospecies 13TU and A. baumannii wherein this plasmid appeared to be acquired via one-ended transposition (Tn6080)23. One study from India has noted very high carbapenem resistance (imipenem MIC -128 μg/ml) among the isolates that bear blaOXA-51 on their chromosome24. In our study, plasmid-borne blaOXA-51 bearing isolates had higher MICs to carbapenems than those bearing blaOXA-51 only on chromosomes. This could be due to increased gene dosage provided by the higher copy number of associated plasmids.
Multicopy blaOXA-58 gene has been a source of high-level carbapenem resistance and plasmid borne blaOXA-58 is apparently the only mechanism of resistance which can be transferred by conjugation142223. Earlier, it was shown that an 11 kb A. baumannii plasmid (pTVICU53) harbouring 13-open reading frames (ORFs) including blaOXA-58 with an upstream insertion of ISAba3 and IS1008 provided two independent promoters for the transcription control of OXA-5825. pTVICU53 transformation was shown to increase the carbapenem MICs by 64 to 256-fold and the deletion of these promoters resulted in drastic reduction of carbapenem MIC. Such findings underscore that A. baumannii might develop carbapenem resistance with a single transformation step, taking up a plasmid containing a genetic construct with a potentially high level of transcription of the OXA-genes25.
In summary, multiple drug resistance in Acinetobacter species appears to be a combined effect of lateral gene transfer (plasmids and integrons) and clonal dissemination of MDR clones. This is likely scenario in any hospital settings including ours. Studies on identification of resistant plasmids and deducing their likely mode of transfer are important to control and eradicate persistent endemic and epidemic clones of A. baumannii. In addition, stringent infection control measures and their implementation are of prime importance that can prevent further propagation of resistance.
Acknowledgment
Authors thank the Indian Council of Medical Research (ICMR), Government of India, New Delhi, for funding the study (No.5/3/3/14/2007-ECD) and University Grants Commission (UGC), Government of India for financial assistance under Special Assistance Programme (SAP) F.3 -21/2009(SAP-II).
References
- The Acinetobacter baumannii oxymoron: commensal hospital dweller turned pan-drug-resistant menace. Front Microbiol. 2012;3:148.
- [Google Scholar]
- Acinetobacter baumannii: emergence of a successful pathogen. Clin Microbiol Rev. 2008;21:538-82.
- [Google Scholar]
- Metallo-beta-lactamases in Gram-negative bacteria: introducing the era of pan-resistance? Int J Antimicrob Agents. 2009;33:405.e1-7.
- [Google Scholar]
- Simplified panel of assimilation tests for identification of Acinetobacter species. Indian J Pathol Microbiol. 2003;46:700-6.
- [Google Scholar]
- Molecular characterization and control of Acinetobacter baumannii isolates resistant to multi-drugs emerging in inter-intensive care units. Ann Clin Microbiol Antimicrob. 2014;13:36.
- [Google Scholar]
- Plasmids and their usefulness in molecular cloning. In: Sambrook J, Russell DW, eds. Molecular cloning: a laboratory manual (3rd ed). New York: Cold Spring Harbor Laboratory Press; 2001. p. :1.1-1.163.
- [Google Scholar]
- Plasmid-borne extended-spectrum beta-lactamase in a clinical isolate of Acinetobacter baumannii. J Med Microbiol. 2003;52:1125-7.
- [Google Scholar]
- Performance standards for antimicrobial susceptibility testing, 21st informational supplement (M100-S22) Wayne, PA: CLSI; 2012.
- [Google Scholar]
- Imipenem-EDTA disk method for differentiation of metallo-β-lactamase-producing clinical isolates of Pseudomonas spp. and Acinetobacter spp. J Clin Microbiol. 2002;40:3798-801.
- [Google Scholar]
- A plasmid-borne blaOXA-58 gene confers imipenem resistance to Acinetobacter baumannii isolates from a Lebanese hospital. Antimicrob Agents Chemother. 2008;52:4115-20.
- [Google Scholar]
- Investigation of a nosocomial outbreak of imipenem-resistant Acinetobacter baumannii producing the OXA-23 beta-lactamase in korea. J Clin Microbiol. 2005;43:2241-5.
- [Google Scholar]
- Phenotypic and genotypic assays for detecting the prevalence of metallo-β-lactamases in clinical isolates of Acinetobacter baumannii from a South Indian tertiary care hospital. J Med Microbiol. 2009;58:430-5.
- [Google Scholar]
- Antibiotic resistance mechanisms in Acinetobacter. In: Towner KJ, Bergogne-Bérézin E, Fewson CA, eds. The biology of Acinetobacter: taxonomy, clinical importance, molecular, biology, physiology, industrial relevance. New York: Plenum press; 1991. p. :83-111.
- [Google Scholar]
- Carbapenem resistance in Acinetobacter baumannii: Mechanisms and epidemiology. Clin Microbiol Infect. 2006;12:826-36.
- [Google Scholar]
- Transferable amikacin resistance in Acinetobacter spp. due to a new type of 3’-aminoglycoside phosphotransferase. Antimicrob Agents Chemother. 1988;32:15-9.
- [Google Scholar]
- Plasmid distribution & antimicrobial susceptibility patterns of Acinetobacter genospecies from healthy skin of a tribal population in western India. Indian J Med Res. 2007;125:79-88.
- [Google Scholar]
- Plasmid-mediated transfer of the bla(NDM-1) gene in Gram-negative rods. FEMS Microbiol Lett. 2011;324:111-6.
- [Google Scholar]
- Distribution of intrinsic plasmid replicase genes and their association with carbapenem-hydrolyzing class D beta-lactamase genes in European clinical isolates of Acinetobacter baumannii. Antimicrob Agents Chemother. 2011;55:2154-9.
- [Google Scholar]
- Role of common blaOXA-24/OXA-40-carrying platforms and plasmids in the spread of OXA-24/OXA-40 among Acinetobacter species clinical isolates. Antimicrob Agents Chemother. 2012;56:3969-72.
- [Google Scholar]
- Acinetobacter radioresistens as a silent source of carbapenem resistance for Acinetobacter spp. Antimicrob Agents Chemother. 2008;52:1252-6.
- [Google Scholar]
- Characterization of pABVA1, a plasmid encoding the OXA-24 carbapenemase from Italian isolates of Acinetobacter baumannii. Antimicrob Agents Chemother. 2009;53:3528-33.
- [Google Scholar]
- Horizontal transfer of the OXA-24 carbapenemase gene via outer membrane vesicles: a new mechanism of dissemination of carbapenem resistance genes in Acinetobacter baumannii. Antimicrob Agents Chemother. 2011;55:3084-90.
- [Google Scholar]
- Emergence and distribution of plasmids bearing the blaOXA-51-like gene with an upstream ISAba1 in carbapenem-resistant Acinetobacter baumannii isolates in Taiwan. Antimicrob Agents Chemother. 2010;54:4575-81.
- [Google Scholar]
- Carbapenem-hydrolyzing oxacillinase in high resistant strains of Acinetobacter baumannii isolated from India. Microb Pathog. 2012;53:81-6.
- [Google Scholar]
- Acquisition of a plasmid-borne blaOXA-58 gene with an upstream IS1008 insertion conferring a high level of carbapenem resistance to Acinetobacter baumannii. Antimicrob Agents Chemother. 2008;52:2573-80.
- [Google Scholar]






