Translate this page into:
Molecular genotyping of ABO blood groups in some population groups from India
Reprint requests: Dr Ajit C. Gorakshakar, National Institute of Immunohaematology (ICMR), 13th Floor New Multistoreyed Building, K.E.M. Hospital Campus, Parel, Mumbai 400 012, India e-mail: ajit5678@yahoo.com
-
Received: ,
This is an open-access article distributed under the terms of the Creative Commons Attribution-Noncommercial-Share Alike 3.0 Unported, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
This article was originally published by Medknow Publications & Media Pvt Ltd and was migrated to Scientific Scholar after the change of Publisher.
Abstract
Background & objectives:
Indian population is characterized by the presence of various castes and tribal groups. Various genetic polymorphisms have been used to differentiate among these groups. Amongst these, the ABO blood group system has been extensively studied. There is no information on molecular genotyping of ABO blood groups from India. Therefore, the main objective of this study was to characterize the common A, B and O alleles by molecular analysis in some Indian population groups.
Methods:
One hundred samples from the mixed population from Mumbai, 101 samples from the Dhodia tribe and 100 samples from the Parsi community were included in this study. Initially, the samples were phenotyped by standard serologic techniques. PCR followed by single strand conformational polymorphsim (SSCP) was used for molecular ABO genotyping. Samples showing atypical SSCP patterns were further analysed by DNA sequencing to characterize rare alleles.
Results:
Seven common ABO alleles with 19 different genotypes were found in the mixed population. The Dhodias showed 12 different ABO genotypes and the Parsis revealed 15 different ABO genotypes with six common ABO alleles identified in each of them. Two rare alleles were also identified.
Interpretation & conclusions:
This study reports the distribution of molecular genotypes of ABO alleles among some population groups from India. Considering the extremely heterogeneous nature of the Indian population, in terms of various genotype markers like blood groups, red cell enzymes, etc., many more ABO alleles are likely to be encountered.
Keywords
ABO blood group system
Dhodia
India
Parsi
SSCP analysis
ABO blood group system is considered as one of the most important blood group systems in transfusion medicine and population genetics. The serological and genetic characteristics of this system have been well established. There are three major alleles viz., A, B and O at the ABO locus. The locus for ABO gene is on chromosome number 9 (9q 34.1 - 34.2)1. The antigenic determinants are oligosaccharides which are located on either glycospingolipids or glycoproteins. These are not direct gene products of the ABO genes. These genes encode enzymes which are collectively known as glycosyltransferases, and these enzymes transfer specific sugar residues to a precursor substance (H antigen) to produce the A and B antigens. A variety of polymorphisms has been reported in the ABO glycosyltransferase gene1. For convenience, A group glycosyltransferase sequence is considered as a reference sequence and compared to it B and O groups are identified.
Initially specific restriction endonucleases were used to identify common ABO alleles2. Later on by using gene scanning techniques like denaturing gradient gel electrophoresis (DGGE), single strand conformation polymorphism (SSCP) followed by DNA sequencing, several alleles have been identified34. Using these techniques, several populations like Caucasians, Orientals, Amerindians, Brazilians, Kuwaitis and Jordanians56789 from different countries have been screened for detection of different ABO alleles.
India is a multi-ethinic country harbouring 4635 ethinic groups and 635 scheduled tribes as per the 2001 census study10. Genetic variation between inter- and intra-populations have been studied extensively in the non tribal as well as tribal populations using various blood groups and biochemical markers1112. The ABO blood group system has been studied on the largest number of Indian populations131415. In general, higher incidence of B group than A group is characteristic of the people of Indian subcontinent1617. Knowledge of distribution of ABO blood groups is important because certain diseases and malignancies have been found to be associated with the ABO blood groups181920. Bandyopadhyay et al21 studied ABO blood groups in couples with spontaneous abortions and compared them with those with normal newborns from the same area and concluded that ABO incompatibility between the couples could be responsible for early spontaneous abortions and heterozygote selection of ABO blood group genotypes.
Some studies have reported the absence of significant association between malaria and ABO blood groups19, while others have shown high frequency of malaria among blood group A individuals as compared with those with other blood groups22. Carvalho et al23 have tried to find the association if any, between various ABO alleles mainly ‘O’ group alleles and falciparum malaria patients from Brazil. They could not detect significant differences in the frequency of individuals with the various alleles between malaria patients and the general population.
There is no report on molecular genotyping of ABO blood groups from India. This preliminary study was carried out to assess allelic variation and the nature of diversity of ABO blood groups at molecular level in some Indian population groups.
Material & Methods
Samples from three population groups were taken for ABO blood group genotyping. Informed consent was obtained from all the individuals included in the study and the protocol was approved by the ethical committee of the National Institute of Immunohaematology (NIIH), Mumbai, India. Blood samples (5 ml) from 100 individuals from mixed population from Mumbai were collected in EDTA by organizing camps during 2005-2009, and some samples referred to National Institute of Immunohaematology for various studies, were also included. The samples of the Dhodia and the Parsis collected from various camps were not included in mixed population.
Dhodias are the third largest tribal group from Gujarat located in Surat, Valsad districts of Gujarat and also in Madhya Pradesh, Maharashtra and Rajasthan. They live in small bamboo huts made with tiled roofs. Their main source of livelihood is agriculture as well as hunting and fishing. They are endogamous practising clan exogamy, but do not practice consanguineous marriage. Blood samples from 101 individuals from Dhodia tribal group were collected from Pardi, Dharampur, Chikhli and Vasda villages of Valsad district in Gujarat.
Parsis are Zorastrians who arrived in India from Persia. They landed in Diu, off the coast of Gujarat in India. There are no sub-castes in this religion. Marriage with outsiders is very rare. Blood samples from 100 individuals from Parsi community were collected from Valsad, Chikhli and Bilimora villages of Valsad District from South Gujarat and as well as from Mumbai.
Blood grouping was performed by standard tube technique24 using monoclonal antiserum obtained from Ortho Diagnostic, USA. The phenotypes A1 and A2 were differentiated by using anti-A1 lectin prepared from Dolichos biflorus seeds indigenously24.
The DNA was extracted from whole blood using standard phenol-chloroform technique25. Initially, PCR - restriction fragment length polymorphism (RFLP) analysis was performed2 on samples from the heterogeneous mixed population for molecular ABO genotyping. The restriction enzymes like Mvn I/BstU I (nt 188/189), Kpn I (nt 261), Nar I/BssH II (nt 526) and Alu I (nt 467 & 703) obtained from New England Biolabs, USA, were used to identify O01 (O1), O03 (O2), O02 (OIV), B101 (B), A201(A2)/A102(AIV) alleles, respectively.
Three sets of specific primers26 (Sigma, USA) were used to amplify three fragments covering exons 6 and 7 of the ABO gene. Taq polymerase enzyme and dNTPs were obtained from Fermentos, USA and Roche, Switzerland, respectively. The fragment 1 (F1) was amplified covering the sequence from exon 6 and its immediate flanking region, the fragment 2 (F2) was amplified covering the sequence from 5’ end of exon 7 and the fragment 3 (F3) was amplified covering the sequence from 3’ end of exon 7 and the 3’ untranslated region. The single nucleotide polymorphism (SNPs) at nt 261 and 297 in fragment 1, at nt 467, 526, 646, 657 and 681 in fragment 2 and at nt 1059, 1061 and 1096 in fragment 3 were the known SNPs in ABO gene. The amplified products were pooled in a single tube and single strand conformational polymorphism (SSCP) was run using 9%T/1%C (T-Total concentration of acrylamide gel and C-concentration of cross linker) poly acrylamide gel electrophoresis (PAGE) at 400V for 2 and 1/2 h as described previously26. The nine SNPs mentioned above produce a particular pattern of SSCP. Initially the samples whose genotypes were identified by PCR-RFLP analysis were run to develop a catalogue of SSCP patterns corresponding to different genotypes either homozygous or heterozygous. The ABO genotypes of Dhodias and Parsis were determined using this catalogue.
After the SSCP run was complete the bands were visualized by silver staining. The SSCP gel was stained by background free silver staining method27. For easy visualization of specific bands sodium thiosulphate was used in staining procedure to make the gel free from yellow or brown background by non-specific deposits of insoluble silver salts27.
The samples which showed altered banding pattern in SSCP were characterized by DNA sequencing. The DNA sequencing was carried out in the ABI Prism 310 Genetic Analyzer (Applied Bio-system, C.A. USA). The resultant data were analysed using Macintosh sequence analysis 3.4 software, USA.
The ‘A’ variant phenotype was identified by SSCP and confirmed by sequence specific PCR (SSP)28 while the ‘O’ variant phenotype, the rare allele was identified by DNA sequencing.
Results
The seven common ABO alleles were characterized by a set of three haplotype-specific SSCP patterns by the three amplified fragments (Figure A). In the mixed population, all the seven alleles were found with a combination of 19 ABO blood group genotypes, in the Dhodia tribe six alleles were found with a combination of 12 ABO blood group genotypes and in the Parsi population six alleles were found with a combination of 15 ABO blood group genotypes. Four homozygous patterns from the known genotypes and the combination of other patterns were utilized to develop the catalogue for SSCP.
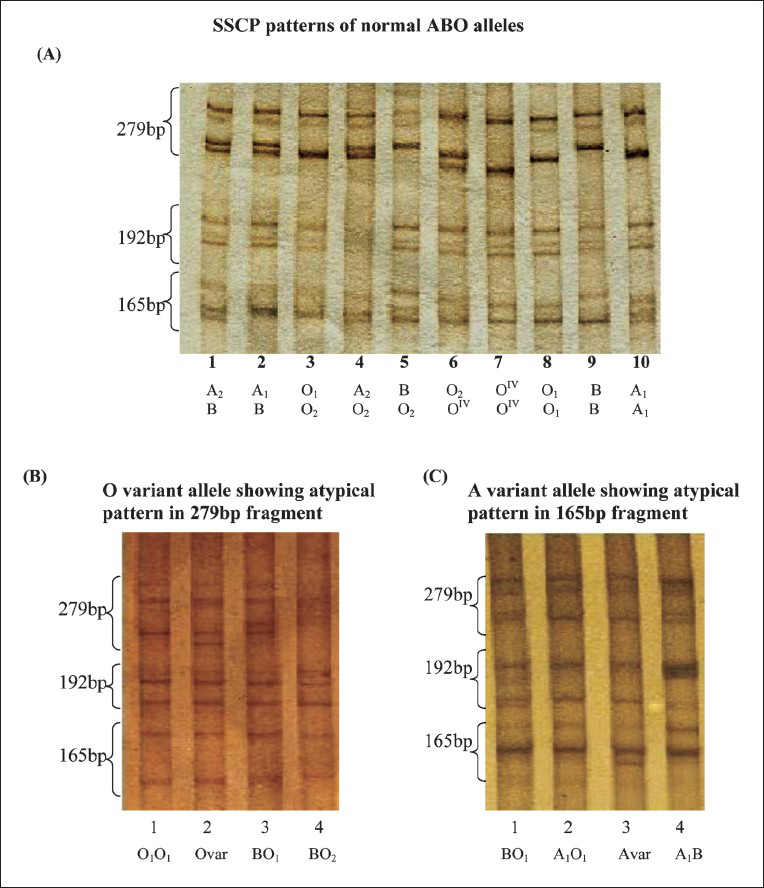
- SSCP patterns of common as well as variant ABO alleles. Fig. (A) shows the single strand conformational polymorphism (SSCP) patterns of different homozygous and heterozygous ABO alleles (Lane1-10: A2B, A1B, O1O2, A2O2, BO2, O2OIV, OIVOIV, O1O1, BB & A1A1). Fig. (B) shows the normal SSCP pattern of ABO alleles (lane 1, 3, 4) Lane 2: Atypical pattern of O variant allele. Fig. (C) shows the normal SSCP pattern of ABO alleles (lane 1, 2, 4) with atypical pattern of A variant allele (lane-3).
In addition to the common alleles, two rare alleles were detected in the mixed population. These showed altered mobility in SSCP banding pattern, both the individuals carrying these alleles were from Sindhi community. The altered SSCP banding pattern was detected in fragment 279 bp (Fig. B). On DNA sequencing the following rare polymorphisms were observed which confirmed the allele as O209 i.e. 542 - G→A, 646 - T→A, 681 - G→A and 771 - C→T. In another variant phenotype, the altered SSCP banding pattern was detected in fragment 165 bp (Fig. C) which was confirmed by PCR-RFLP and sequence specific PCR (SSP). This variant phenotype did not show the C→T polymorphism at nucleotide position 467 as seen in cases of A201 (A2) and A102 (AIV) but it showed the (-C) at the 3’ terminal of exon seven at nucleotide position 1059 and therefore, named as A206.
The B101 (B) allele was more common in mixed population as well as in Parsis followed by O01 (O1) and A101 (A1) alleles, whereas A101 (A1) allele was more common in Dhodia tribe followed by O01 (O1) and B101 (B) alleles (Table I). The SSCP pattern of O01 (O1) allele differed from A101 (A1) allele only in the fragment-1 (192 bp), the SSCP pattern of A201 (A2) allele differed from A101 (A1) allele only in the fragment-3 (165 bp) and the B101 (B) and O03 (O2) alleles showed the same SSCP pattern except in the fragment-2 (279 bp). The O02 (OIV) allele had a different SSCP banding pattern in both fragment-1 (192 bp) and fragment-2 (279 bp).

Distribution of the ABO genotypes in the three groups revealed that the ‘A101O01’ (A1O1) genotype was common in the mixed population (0.11) and Dhodias (0.217), but among the Parsi population the ‘A101O02’ (A1OIV) genotype was more prevalent (0.1) than other genotypes within ‘A’ blood group. In ‘B’ blood group the ‘B101O01’ (BO1) genotype was more prevalent than other genotypes in all the three populations. The ‘O01O01’ (O1O1) and the ‘O01O02’ (O1O1IV) genotypes were the most common genotypes in O blood group in all the three groups studied. Similarly, the O01 (O1) and O02 (OIV) alleles were found to be common in the population groups and the non-deletional O03 (O2) allele was rare in our population groups.
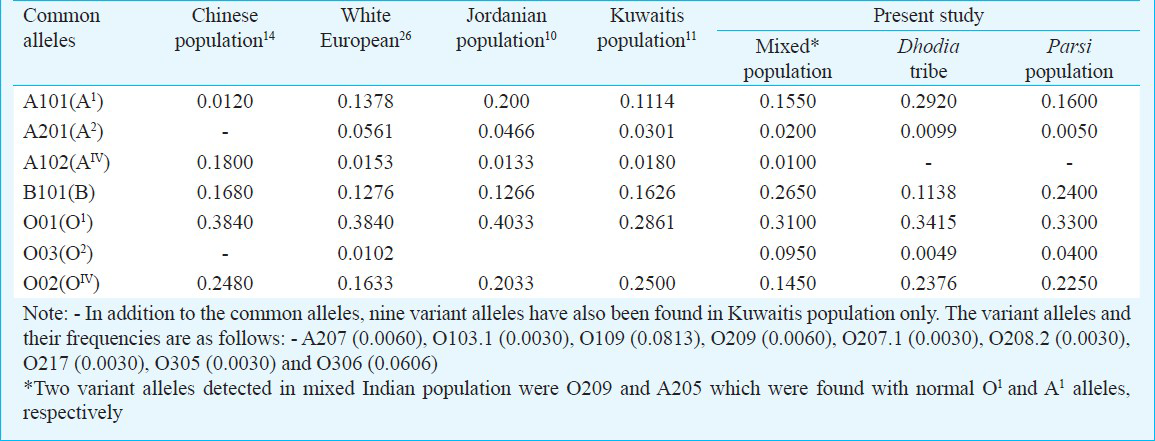
Table II shows the comparison of ABO allele frequencies in different world populations with the present study in Indian population. Seven common alleles [A101, (A1), A102 (AIV), A201 (A2), B101 (B), O01 (O1), O02 (OIV) & O03 (O2)] were identified in the mixed population, while A102 (AIV) allele was absent in Dhodias and Parsis. Absence of A102 (AIV) allele and presence of only one sample with A201 (A2) allele suggest homogeneity if ‘A’ alleles in A group among Dhodias. Only A101 (A1) within ‘A’ group was seen in this group. They showed a high frequency of A101 (A1) allele (0.2920) as compared to the mixed population (0.1550) and Parsis (0.1600), while frequency of B allele was lowest (0.1138) in Dhodias.
Discussion
Cloning of ABO genes and elucidation of the molecular basis of its major alleles had helped us to directly determine the ABO genotypes without family study. Three main alleles (A, B & O) were identified by restriction enzyme analysis2.
Using the SSCP technique as many as 13 different common and rare alleles were identified by using PCR products from exons 6 and 729. The same approach was further developed by multiplexing three PCRs in a single tube and analyzing the products by SSCP in a single lane26. This protocol can identify base changes at nine positions (261, 297, 467, 526, 646, 657, 681, 1059 to 1061, and 1096) and can identify the seven common alleles. In the present study, this same protocol was adopted.
The three population groups including mixed population showed higher percentage of A101 (A1) allele over the A102 (AIV) and A201 (A2) alleles. The results were similar to the Caucasian, white European and Kuwaiti population2. The A102 (AIV) allele was found to be frequent than the A101 (A1) allele in Chinese population while in Japanese and German population30, it was less frequent than A101(A1) allele. In the present study, it was seen with low frequency in the mixed population group only.
Molecular analysis of ‘O’ alleles showed heterogeneity. Johnson & Hopkinson3 studied 50 ‘O’ group individuals by denaturating gradient gel electrophoresis (DGGE) and found four different alleles which they named as O1, O2, O3 and O4. Olsson et al5 identified several rare alleles especially among blacks from Brazil. Roubinet et al9 studied O alleles in five ethnic groups from Ivory Coast, Equator and Bolivia. Amongst these 13 ‘O’ alleles were found among Akans from Ivory Coast, while Yang et al8 found eight O alleles in Chinese population. As compared to this, two deletional O group alleles [O01 (O1) & O02 (OIV)] were detected in all the three groups in the present study. A non deletional O group allele, O03 (O2) was seen with very low frequency (0.005 - 0.0950) in all the groups, while only one case of O209 or O11 was detected.
The frequency distribution of different ABO alleles in different world populations is compared with the Indian population groups screened in the present study. A201 (A2) allele is absent in Chinese population while A102 (AIV) allele was not seen in Dhodias and Parsis from India. The allele O03 (O2) is extremely rare or does not exist in Orientals like Chinese, Japanese, Koreans, etc1. This allele is also not found in Kuwaitis7 and Jordanians6. However, it has been found in low frequencies (0.7-2.8%) in various Caucasian groups like Germans, Danish and in blacks1.
The two rare alleles detected in the Sindhis from general population are O11 (O209) and A206. The O11 (O209) accounts for 4 per cent of all O alleles in Cayapas from Equador, 12 per cent in Aymaras from Bolivia and interestingly 43 per cent in Amerindians from Brazil9. Two A206 alleles were earlier found in Caucasian population1. Considering the diversity of the Indian population duet to a large number of ethnic group one can expect to get many more alleles when more and more population groups will be screened.
Molecular genotyping of ABO blood groups is a useful tool in forensic science and paternity testing. In a country like India, where a large number of endogamous ethnic groups are there, this technique will help to understand the heterogeneity at ABO locus within and between the groups and to prepare database of these alleles in the Indian population.
Using molecular genotyping techniques by SNP analysis, several inactive O alleles have been identified by Yip1, which has helped us to understand the process of inactivation of O alleles. Similarly, study of various ABO alleles has helped us to understand rare and intriguing phenotypes like B(A) or cis AB. The occurrence of hybrid alleles explains abnormal inheritance in some pedigrees.
In conclusion, the present study reports detailed distribution of ABO alleles and genotypes among Parsis, Dhodias and mixed population from Mumbai, India. Using multiplex PCR-SSCP method 21 molecular genotypes formed by six ABO alleles weree identified in these population groups directly without family studies. These findings initiate the formation of a database of ABO alleles in the Indian population. Considering the large number of ethnic groups present in our country considerable heterogeneity is likely to be seen at ABO locus. This can be used as a marker for anthropological studies in India to study the origins and movements of populations.
Acknowledgment
Authors acknowledge the staff of Valsad Raktadan Kendra for providing the blood samples of Dhodias and Parsi populations by organizing camps in various villages.
References
- ABO genotyping by polymerase chain reaction-restriction fragment length polymorphism. Immunohematology. 1996;12:143-8.
- [Google Scholar]
- Detection of ABO blood group polymorphism by denaturing gradient gel electrophoresis. Hum Mol Genet. 1992;1:341-4.
- [Google Scholar]
- Molecular genetic analysis of variant phenotypes of the ABO blood group system. Blood. 1996;88:2732-7.
- [Google Scholar]
- Heterogeneity of the O alleles at the blood group ABO locus in Amerindians. Vox Sang. 1998;74:46-50.
- [Google Scholar]
- Phenotype prediction by DNA-based typing of clinically significant blood group systems in Jordanian blood donors. Vox Sang. 2002;83:55-62.
- [Google Scholar]
- ABO blood group in Kuwaitis: detailed allele frequency distribution and identification of novel alleles. Transfusion. 2006;46:773-9.
- [Google Scholar]
- Molecular polymorphism of O alleles in the Chinese Han population. Ann Clin Lab Sci. 2007;37:71-4.
- [Google Scholar]
- Molecular polymorphism of O alleles in five populations of different ethnic origins. Immunogenetics. 2001;53:95-104.
- [Google Scholar]
- Genetic polymorphisms in human populations in India. In: Satyavati GV, ed. People of India - Some genetical aspects. New Delhi: ICMR; 1983. p. :1-29.
- [Google Scholar]
- Genetic atlas of Indian tribes. Mumbai: Institute of Immunohaematology (ICMR); 1986.
- [Google Scholar]
- Distribution of ABO blood groups on the Indian subcontinent: a cluster analytic approach. Curr Anthropol. 1982;23:539-66.
- [Google Scholar]
- Genetic studies in Ratnagiri and Sindhudurg districts of Maharashtra. Incidence of ABO. Rho (D) Ina antigens, G6PD deficiency and abnormal haemoglobins. J Indian Antpropol Soc. 1987;22:38-46.
- [Google Scholar]
- ABO and Rh blood group distribution among Kunbis (Maratha) population of Amravati District, Maharashtra-India. Asiatic J Biotechnol Resour. 2011;2:479-83.
- [Google Scholar]
- A study on blood groups and serum proteins in Bengalee populations of Calcutta, India. Anthropol Anz. 1994;52:215-9.
- [Google Scholar]
- Genetic markers in human blood. In: Bhasin V, Bhasin MK, eds. Anthropology today (trends, scope & application). New Delhi: Kamala-Raj Enterprises; 2007. p. :247-347.
- [Google Scholar]
- Frequency of ABO blood groups among the diabetes mellitus type 2 patients. J Coll Physicians Surg Pak. 2003;13:453-5.
- [Google Scholar]
- The ABO blood group system and Plasmodium falciparum malaria. Blood. 2007;110:2250-8.
- [Google Scholar]
- Maternal fetal interaction in the ABO system: a comparative analysis of healthy mother and couples with spontaneous abortion in Bengalee population. Am J Hum Biol. 2011;23:76-9.
- [Google Scholar]
- Association of ABO groups in malaria infection of variable severity. J Vector Borne Dis. 2012;49:78-81.
- [Google Scholar]
- Frequency of ABO blood group system polymorphisms in Plasmodium falciparum malaria patients and blood donors from the Brazilian Amazon region. Genet Mol Res. 2010;9:1443-9.
- [Google Scholar]
- Techniques in Blood Banking. Mumbai, New Delhi: Institute of Immunohaematology, ICMR; 1985.
- [Google Scholar]
- Laboratory procedures for DNA analysis. WHO training course in standard techniques and advanced methodologies for the control of hereditary anemias. In: Herakleion 1987. 1987. p. :5.
- [Google Scholar]
- Single-tube multiplex PCR-SSCP analysis distinguishes 7 common ABO alleles and readily identifies new alleles. Blood. 2000;95:1487-92.
- [Google Scholar]
- Improved silver staining protocols for high sensitivity protein identification using matrix-assisted laser desorption/ionization-time to flight analysis. Proteomics. 2001;1:1359-63.
- [Google Scholar]
- Polymorphisms at the ABO locus in subgroup A individuals. Transfusion. 1996;36:309-13.
- [Google Scholar]
- Extensive polymorphism of ABO blood group gene: three major lineages of the alleles for the common ABO phenotypes. Hum Genet. 1996;97:777-83.
- [Google Scholar]
- Genotyping of ABO blood group system: analysis of nucleotide position 802 by PCR-RFLP and the distribution of ABO genotypes in a German population. Intl J Legal Med. 1996;109:90-3.
- [Google Scholar]
- Heterogeneity of the blood group Ax allele: genetic recombination of common alleles can result in the Ax phenotype. Transfus Med. 1998;8:231-8.
- [Google Scholar]






