Translate this page into:
Molecular epidemiology of human papillomavirus variants in cervical cancer in India
For correspondence: Dr Manpreet Kaur, Department of Human Genetics, Guru Nanak Dev University, Amritsar 143 005, Punjab, India e mail: dr.manpreetdhuna@gmail.com
-
Received: ,
Accepted: ,
Abstract
Background & objectives
Cervical cancer (CC) has been documented as the fourth most common cancer worldwide. Persistent infections with high-risk human papillomavirus (hr-HPV) have been suggested in the development of CC. Although prophylactic vaccines are available for the prevention of prevalent hr-HPV types, intra-type variations exist within a particular HPV type that has varying oncogenic potential as well as the mechanism of pathogenicity and varying neutralization by antibodies. Therefore, we carried out a systematic review to determine the distribution of HPV intra-typic variations in different geographical locations of India and their reported implications.
Methods
Preferred Reporting Items for Systematic Reviews and Meta-Analysis (PRISMA) guidelines were followed to retrieve relevant articles from the standard databases using appropriate keywords. Consequently, 17 articles were included in the current review after screening based on inclusion and exclusion criteria.
Results
The majority of articles included in this review reported variations within the HPV16 E6 gene, followed by the L1 and E7 genes. Analysis of available data indicated the differential regional distribution of some variations. These variations have also been reported to impact the biological functions of various viral proteins.
Interpretation & conclusions
The distribution of lineages varied with the different genomic regions sequenced. Additionally, there were certain unique and common variations in the HPV genome with respect to geographical regions. Hence, we suggest the identification of region-specific variations for the development of diagnostic and prognostic interventions.
Keywords
Asian American variants
European variants
HPV
intratypic variants
lineage
sublineage
variations
350G variant
Cervical cancer (CC) is the fourth most common cancer and the fourth leading cause of cancer deaths among women worldwide1. The incidence of CC is disproportionately high among low- and middle-income countries. In India, it is the third most frequent cause of cancer mortality2. Persistent infections with high-risk human papillomavirus (hr-HPV) have been documented as a major cause of CC and its involvement in other types of cancers3. HPV is a circular, non-enveloped, double-stranded DNA virus infecting basal keratinocytes of the host4. HPV genome can be divided into three regions, namely, (i) early gene region (E) consisting of E1, E2, E4, E5, E6, and E7, (ii) late gene region (L) including L1 and L2 genes, and (iii) non-coding, long control region (LCR). In addition, a smaller non-coding region between E5 and L2 genes is referred to as non-coding region 2 (NCR2). The important functions of these regions are given in table I5-7. These variations have also been suggested to modulate their oncogenic potential and may also facilitate immune evasion8,9. HPV can be classified into various types depending upon at least 10 per cent difference in its highly conserved L1 gene10. Furthermore, nucleotide variations within HPV types can be used to classify each type into variant lineages and sub-lineages11,12. Lately, the HPV 16 complete genome has been classified into 4 lineages, named A, B, C, and D, which are further divided into sub-lineages, A1-3 (European), A4 (Asian), B1-4 (African1), C1-4 (African2), D1 (North American), D2-3 (Asian American) and D413. Different geographical prevalences of certain intra-type variants have been reported8. Consequently, intra-type variants differ in their oncogenic potential and mechanism of oncogenicity14, as Asian American variants of HPV16 have been reported to retain the E2 gene, suggesting an alternative mechanism to regulate E6 and E7 transcription15. Studies have also reported reduced sensitivities of HPV variants to cross-neutralization by vaccine antibodies and/or mAb raised against A antigens16-20.
| Viral genomic regions | Properties | Biological functions |
|---|---|---|
| L1 | The L1 proteins are mainly expressed during the later stages of viral infections. It consists of variable & constant regions, where the variable region carries surface-specific antigenic epitopes that cause the production of neutral antibodies |
The major capsid protein (L1) forms the pentameric monomers that comprise 5L1 and 1L2 protein, which ultimately forms 72 capsomeres that cover the virus. It can produce HPV-specific antibodies due to the presence of multiple surface epitopes |
| L2 | Minor capsid protein along with L1 forms the capsid of the virus | It is responsible for interaction with host receptors & facilitates endocytosis of the viral genome |
| E1 | It is the highly conserved protein among HPV types and is expressed during early infection | It is responsible for viral replication. It also regulates epigenetic modulations within the cell as well as is also reported in the immune response |
| E2 | It consists of two domains, the N terminus conserved domain, involved in transactivation and DNA replication, and the C terminus DNA binding domain that facilitates dimerization. | The E2 protein along with the E1 protein is involved in viral DNA replication and transcription. It is also involved in the repression of E6 and E7 gene expression by binding to its promotor within the LCR |
| E4 | The E4 gene is present within the E2 gene and is translated as E1ˆE4 fusion protein. It consists of a leucine cluster motif in its N terminus that aids in association with keratin. | The E4 protein is involved in viral release and transmission |
| E5 | It is a membrane-bound protein that consists of 3 domains | It is involved in hyperproliferation and cancer progression. It also induces angiogenesis, suppresses tumour suppressor proteins, and promotes anti-apoptosis |
| E6 | E6 protein consists of 2 zinc finger domains, flanked by 4cys-X-X-cys motifs, as well as a PDZ-binding motif | It is mainly involved in p53 degradation, to evade cell death, proliferation, cell invasion, immune evasion, and immortalization |
| E7 | E7 protein consists of three conserved regions. The conserved region near the C-terminus encodes the zinc finger domain flanked by 2CXXC motifs | The E7 protein is involved in inactivation of pRB and the downregulation of E2F. it also causes cell cycle progression, cell proliferation, cell invasion, and inflammation |
| LCR | LCR can be subdivided into 3 segments namely the 5’ segment, central segment, and 3’ segment, which are sites for binding to viral and host factors | LCR is responsible for viral replication, regulation of viral transcription, and binding to various viral and host factors |
LCR, long control region; HPV, human papillomavirus
Therefore, the identification of HPV variants may be vital for the development of new diagnostic and therapeutic interventions. Hence, we carried out the current review to determine the distribution of HPV variants within the Indian population and their reported implications, which can subsequently be instrumental in designing population-specific interventions.
Materials & Methods
The study followed the Preferred Reporting Items for Systematic Reviews and Meta-Analysis (PRISMA) guidelines. Articles were retrieved from PubMed, Scopus, Science Direct, and Web of Science on September 9, 2023, and again on November 4, 2023. Key words used for assessing relevant literature included ‘HPV variants in cervical cancer’, ‘nucleotide variations AND HPV AND cervical cancer’, ‘single nucleotide polymorphisms AND HPV AND cervical cancer’, and ‘Human papillomavirus lineages AND cervical cancer’. Grey literature sources like Google Scholar were also consulted to ensure that all pertinent articles were included. A total of 333 articles were obtained after removing duplicates through titles. The remaining articles (n=148) were then screened through their abstracts, and a total of 62 articles were obtained, the full text of which were studied further to exclude irrelevant articles. Finally, 17 relevant articles pertaining to the objective of the current systematic review were included (Fig. 1).
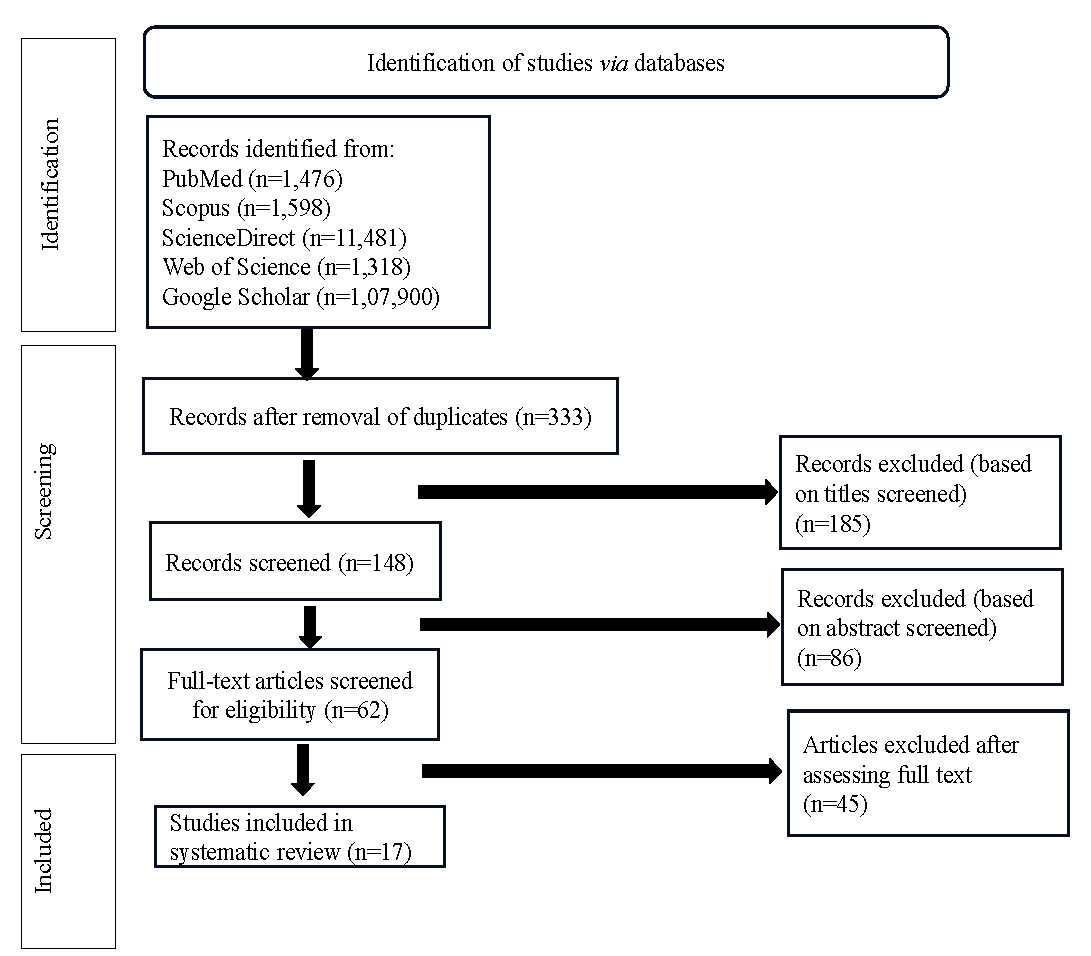
- PRISMA flow diagram for the study selection process.
Inclusion criteria
The inclusion criteria comprised research articles involving participants diagnosed with abnormal cytology or with CC, were exclusively from India, availability of full text, articles in the English language, and those that were relevant as per the objective of the current review.
Exclusion criteria
Exclusion criteria were articles in which participants were coinfected with other pathogens, studied in countries other than India, different diseases, or in women with normal cytology, and articles in which the country of origin of samples could not be determined through text.
Quality Assessment
The methodological evaluation of the given studies was based on the Quality assessment of observational studies21,22. Depending on the study design of the included articles, a total of 10 items were selected for cohort studies, while 12 items were selected for case-control studies. The observational cohort studies with a score of >7 were regarded as high-quality studies, with a score of 4 to 7 as medium quality and below 4 as low-quality studies. Similarly, the observational case-control studies with scores >8 were regarded as high-quality studies, with scores 4 to 8 as medium and below 4 as low-quality studies. The evaluation of the studies was done through discussion, and the results of methodological quality assessment are provided in supplementary tables I and II.
Distribution of studies into different geographical regions
According to geographical location, the relevant studies were segregated into eastern, western, northern, southern, and central regions, and studies involving participants from multiple locations in India. Details of the participants recruited, and the HPV genome region studied have been compiled in table II.
| Regions | Study | Yr | Sample size | HPV | Gene | |
|---|---|---|---|---|---|---|
| East India | West Bengal | To determine the variations within the E2 and LCR regions | 200623 | 49 ISCC cases and 23 controls | 16 | E2 |
| 42 ISCC cases and 19 controls | 16 | LCR | ||||
| West Bengal | To determine distinctive features of HPV16 lineages wrt nucleotide variations, viral load and E7 gene expression | 202224 | 145 ISCC and 24 controls | 16 | Whole genome | |
| West India | Mumbai | To determine the prevalence of molecular variants and their impact on disease outcome | 201425 | 62 cervical cancer patients | 16 | E6 |
| North India | New Delhi, Aligarh | To analyze variations within the viral genome along with its association with age, histopathological grade, and oncogenic potential | 200827 | 31 cervical cancer patients | 16 | L1 |
| 60 cervical cancer patients | 16 | E6 | ||||
| 60 cervical cancer patients | 16 | E7 | ||||
| 60 cervical cancer patients | 16 | LCR | ||||
| North India | To determine the predominance of HPV genotypes, variant classes of hr-HPVs, and viral load among various sample groups | 200928 |
74 ICC 9 SIL 9 cytologically normal controls |
16/18 | E6/E7 | |
| Delhi | To determine gene variants and predict epitopes for common MHC I and MHC II alleles present | 201530 | 221 cervical cancer cases | 16 | E6 | |
| South India | Thiruvananthapuram | To examine the association of variations in HPV oncogenes with disease status | 200231 |
111 patients with HSIL and ICC 32 controls with benign or LSIL |
16 | E6/E7 |
| Vellore | To determine the presence of specific variations within the HPV gene, that have been reported to be significant for biological and immunological functions | 200432 | 76 ICC patients and 16 CIN | 16 | E2 | |
| Karnataka | To determine the prevalence of HPV types, and of different variants of HPV from rural women | 201533 |
15 normal 24 SIL 51 malignant |
16 | L1 | |
| Central India | Chattisgarh | To analyze variations in the HPV oncogenes as well as their effect on protein structure | 202134 | 21 cervical cancer patients | 16 | E6, E7 |
| Multiregional | South and East India | To determine the frequency of the particular HPV variant among women | 200535 |
50 CIN and ICC patients 20 controls |
16 | E6 |
| West Bengal, Chattisgarh, Manipur and Sikkim | To identify sequence variations within the HPV genome and their effects on oncogenicity, disease progression, and pathogenicity | 200836 | 53 ISCC patients and 21 controls | 16 | Whole genome except E1 | |
| Delhi, Kolkata, Bangalore, Mumbai, Thiruvananthapuram, Vellore | To determine gene variations among cervical neoplasia patients | 200937 | 412 SCC | 16 | E6, E7 and L1 | |
| West Bengal, Chattisgarh, Manipur and Sikkim | To extend the above findings by Bhattacharjee26, variations within the entire HPV16 genome were assessed in the episomal HPV genome. In addition, the association of synonymous variations with pathogenesis as well as genes comprising such variations were identified and the significance of variations within non-coding regions was also evaluated. | 201339 |
70 ISCC cases with intact E2 29 controls |
16 | E1 | |
| Pune, Belgaum, Chennai | To characterize HPV gene variants and lineage/sub-lineage distribution and its correlation with sequence, structure, and functional implications. | 202040 |
133 women undergoing screening Normal-43 LSIL-45 HSIL-45 |
16 | L1 | |
| Pune, Belgaum, Chennai | To determine variations within the HPV genome and its associations with different grades of neoplasia as well as the impact of these variations on the regulation of viral transcription. | 202241 |
54 normal 50 LSIL 59 HSIL (as per assigned lineage) |
16 | LCR | |
| Retrieved samples from Pune, Belgaum and Chennai | To identify variations within the HPV gene and its association with disease. Additionally, B- and T-cell epitopes in L2 were identified and the impact of found variations within epitopes was also determined. | 202242 | 41 normal, 45 LSIL, and 62 HSIL | 16 | L2 | |
ISCC, invasive squamous cell carcinoma; ICC, invasive cervical cancer; SCC, squamous cell carcinoma; SIL, squamous intraepithelial lesion; LSIL, low grade SIL; HSIL, high grade SIL; CIN, cervical intraepithelial neoplasia
Results
Quality assessment of the studies
The quality assessment of the included studies found that seven of ten cohort studies were of high quality, and three were of medium quality. Similarly, all the case-control studies were of high quality.
Distribution of HPV variants among various geographical regions
Out of the total studies, most reported HPV16 variants, while a single study reported HPV18 variants. The gene studied the most was the E6 gene, followed by L1 and E7, which had five studies each. However, no study to date reports the epidemiology of variants of other prevalent HPV types in the Indian population. Distribution of nucleotide variations as well as lineages of HPV16, reported in the included studies, have been compiled in table III and IV, respectively. Furthermore, the nucleotide variations and lineage distribution of HPV18 have been compiled in table V.
| Genomic regions | Nucleotide variations/amino acid substitutions | References | |
|---|---|---|---|
| Synonymous variations | Non-synonymous variations (amino acid substitutions) | ||
| East India | |||
| L1 | G5696A, C5756G, A5816T, T5855C, T5909C, A6023T, 6028delA, 6049insA, 6092delA, 6105delA, T6245C, A6314G, C6539A, *C6557T, A6559T, 6560delT, 6561delA, 6606insT, 6626insT, 6638delT, *A6665C, 6669delT, G6719A, A6779T, C6824T, *C6852T, 6857delA, C6863T, 6871delG, 6909delA, 6918delC, 6934delC, T6966C, C6968T, C6969T, T6972C, G6978A, A6983C, 6988delT, 6989delA, 6990delA, *G6992A, A6994G, T6999G, G7008A, 7011delC, C7017A, 7027delT, 7049delA, G7058A, G7062C, A7073T, 7075delT, 7076delT, 7077delA, 7098delA, C7106A, T7130G, 7135delG, G7144C | *C5562G (Q2E), A5620G (Y21C), T5629C (F24S), T5638A (M27K), A5797C (K80T), G5829A (V91l), *C5862T (H102Y), *C6163A (T202N), G6171A (A205T), A6178C (N207T), C6240G (H228D), A6432G (T292A), A6445C (N296T), C6564A (P336T), G6652C (S365T), *A6693C (T379P), A6801T (T415S), 6901insAC (448insS), 6950delGT (465delD), G7033C (R492P), *G7058T (L500F), G7090A (R511Q), A7132C (K525T) | 37 |
| L1 | T5675C, *G5698A, A5755C, *T6247C, A6316G, T6319C C6370T, A6391G, A6425C, C6541A, *A6565G, *C6559T, *A6667C, *G6721A, C6826T, *C6854T, *C6865T, T6889G, A6952G, *C6970T, *G6994A, T7027G, G7032A, A7072T, T7078C, A7141C | *C5864T (H76Y), *C6165A (T176N), G6173A (A179T), *A6180C (N181T), C6182G (P182A), *A6434G (T266A), T6438G (V267G), A6445C (N296T), G6458A (D274N), G6582A (R315Q), T6689G (S351A), *A6695C (T353P), *A6803T(T389S), T7028G (L464V), A7148C (K504Q) | 24 |
| L2 | A4242G, A4266C, *T4281C, A4299C, *A4410G, *G4428T, T4446G, *T4452C, *G4461A, A4467C, A4468C, C4525T, T4545G, *A4599C, A4626T, A4632C, *T4644A, T4647G, T4695C, *C4725T, A4788C, A4887G, G4938A, A4944G, *A4950G, *A5034T, A5073T, G5142A, T5286A, *G5379A, A5409G, T5412G, T5475A, T5478C, *C5487T, A5517G, T5523C, A5532G, A5622G | C4253T (S6F), *T4600C (S122P), G4621A (D129N), G4654A (D140N), A4693C (T153P), A4811C (N192T), A4820G (E195S), *A4969G (T245A), G5008A (E258K), A5027C (N264T), T5041C (S269P), C5042A (S269Y), A5048C (N271H), T5060A (I275N), A5063C (N276T), A5152C (I306L), A5250C (E338D), A5256T (E340D), A5356C (I374L), T5357G (I374S), C5369T (S378V), *T5386G (S384A), *G5389A (V385I), A5466C (L410F), A5492C (I418T), *T5495C (I420T), A5505C (Q423H), *G5506A (A424T), A5518C (I428L), *T5368G (S378V), *C5564G (A443G), G5609A (R458Q) | 24 |
| L2 UTR/NCR2 | A4150C, *C4151A, *A4157G, *T4158A, *C4164T, A4184T, T4186A, T4222C, T4231G, A4233C, A4234C | 24 | |
| E1 | *T921C, T1221C, T1446C, G1941A, T2301C, T2343C, *C2344T, T2470C, T2478C, T2778C | C874T (P4S), *G1293T (Q143H), *T1297A (L145I), *G1363A (G167S), *T1421C (I186T), *A1842G (I326M), G2337A (M491I), *G2650A (E596K) | 24 |
| E2 | *C3007T, *G3058A, A3187C, *A3253G, A3538C, T3694A | C2988T (T78M), A3025C (E90D), C3161T (H136Y), *C3236G (H161D), *A3605G (Q286D), T3517A/C (T254K/N), T3566G (F271V), C3684A (T310K), A3181C (E142D), G3182A (A143T), T3224A (L157I), G3249A (R165Q), C3516A (T254K) | 23 |
| E2 | T2914C, C3007T, G3058A, T3172G, *A3181C, A3187C, A3253G, T3259C, T3313G, T3664C, *C3684A, *T3694A, *T3706C, *C3787A, C3800G, *T3805G, T3820C, *T3847C | C2988T (T78M), C3161T (H136Y), *C3159A(T135K), T3172G (I139M), *G3182A (A143T), A3208C (Q151H), *T3224A (L157I), C3236G (H161D), *G3249A (R165Q), A3285C (K177T), G3778T/C (W341C/W341C) | 24 |
| E2/E4 | A3362G, *A3366C, T3371C, C3377G, T3387C, *C3410T, G3413A, A3605G | *A3362G (N203D/A7A), A3366C (E204A/K9Q), T3371C (S206P), *C3377G (P208A/L12L), T3387C (I211T), *C3410T (P219S), G3413A (A220T), *C3516A (T254N/L59T), *T3517C (T254N/L59T), *A3538C (S261S/Q66P), *T3566G (F271V/H75Q), C3576T (S274L/ H79Y), G3585C (G277A/D82H), A3605G (N284D) | 24 |
| E6 | *T286A, *A289G, *A532G | *G145T (Q14D), *C335T (H78Y), *T350G (L83V), G176A (D25N), G188C (E29Q) | 24 |
| E6 | A181C, T286A, A289G, G522DEL, A532G, C537DEL | G145T (Q14H), G176A (D25N), G188C (E29Q), T311A (Y70N),*C335T (H78Y), A336G (H78C),*T350G (L83V), C506G (R135G) | 37 |
| E7 | G666A, T687C, *T789C, *T795G | *T732C (F57V), C790T (R76C) | 24 |
| E7 | G666A, T687C, T730G, *T789C, *T795G | *T732C (F57V), G823A (G88R), T844DEL (Frameshift), G849C (Q98H), A852DEL (Frameshift, K97N, P98H) | 37 |
| LCR | *T7450C, C7394T, C7395T, A7482G, A7485C, G7489A, Int-del T 7497, C7689A, T7743G, A7729C, C7764T, C7786T, G7868A, G7521A, A7550G, T7568G, C7669T, G7703T, T7713G, T7714G, T7775A, G7826A | 23 | |
| LCR |
A7168G, T7176G, T7187C, *G7193T, A7227C, A7233C, A7376T, C7394T, C7395T, T7401C, G7427A, G7435A, *T7450C, A7482G, A7483C, *A7485C, *G7489A, *G7521A, A7550G, C7577T, C7655A, *C7669T, *C7689A, T7713G *A7729C, *T7743G, *C7764T, A7773C, T7775A, *C7786T, C7792T, G7826A, T7827A, G7834T, G7868A, C7886G, C13T, C24T, A93T, A98C |
24 | |
| West India | |||
| L1 | T5673C, G5696A, T5855C, T5909C, 6020delC, A6023T, 6105delA, T6245C, A6314G, C6265T, A6389G, C6539A, 6554delA, *C6557T, *A6665C, 6706delA, G6719A, *C6852T, T6860C, C6863T, A6935T, A6947G, C6968T, A6983C, *G6992A, T7025G, G7058A, T7076C, A7154G | *C5562G (Q2E), C5600A (Y14X), C5609A (D17E), *C5862T (H102Y), A6021G (T155A), G6024A (E156K), *C6163A (T202N), G6171A (A205T), A6178C (N207T), C6240G (H228D), G6261T (D235Y), C6388T (R277L), A6432G (T292A), G6495T (A313S), 6563delA (K335X), *A6693C (T379P), A6801T (T415S), G6879A (E441K), 6901insAC (448insS), G6945A (E463K), 6950delGT (465delD), G7008C (D484H), *G7058T (L500F), A7087C (K510T), G7090A (R511Q), T7150A (L531Q), A7154C (X532Y) | 37 |
| E6 | A187G | *T350G (L83V), *350T, T310G (F69L), A512T (M137L) | 25 |
| E6 | A111G, T286A, A289G, A320DEL, INS342G, G522T, A529G, T530A, A532DEL, A532C, A532G, A562DEL | A131G (R10G), G145T (Q14H), C158A (L19M), G176A (D25N), A182T (I27L), C315G (S71C), *C335T (H78Y), *T350G (L83V), C479T (H126Y), T521C (C140R), T521G (C140V), INS524A (R141K), C531T (S143I) | 37 |
| E7 | *T789C, *T795G | A619T (T20S), *T732C (F57V), A746DEL (Frame shift), G829DEL (V90C, C91A), T844DEL (Frame shift) | 37 |
| North India | |||
| L1 | A6667C,A6691G, A6721G, C6854T, T6889G, C6970T, G6994A, C6865T | *A6695C (T353P), A6803T(T389S), A6964C, C6906T (S423F), A6924C (Q429P) | 27 |
| L1 | T5673C, T5681C, G5696A, A5795G, A5813G, A5816T, A5834G, T5909C, C5967T, A6023T, A6068G, A6182C, T6245C, G6278A, C6365T, A6389G, A6452G, A6518G, C6539A, *C6557T, *A6665C, G6719A, A6779T, *C6852T, C6863T, T6887G, G6936A, C6968T, *G6992A, T7076C, A7094G | *C5562G (Q2E), A5620G (Y21C), *C5862T (H102Y), G5871T (D105Y), *C6163A (T202N), A6178C (N207T), C6240G (H228D), C6388T (R277L), A6432G (T292A), G6495T (A313S), T6573A (L338l), T6634A (V359D), *A6693C (T379P), A6801T (T415S), G6879A (E441K), 6901insAC (448insS), 6950delGT` (465delD), G7008C (D484H), *G7058T (L500F), G7090A (R511Q) | 37 |
| E6 | T286A, A289G, A532G | G145T (Q14D), C335T (H78Y), *T350G (L83V), T527A (S142T) | 27 |
| E6 | T286A, A289G. A532G | T109C, A131G (R10G), G145T (Q14D), A169C (T-P), G176A (D25N), T178G (D25E), G293A (D-N), C335T (H78Y), T350G (L83V), 403, G507A (R-E), G525T (R-I) | 28 |
| E6 | T286A, A289G | G145T (Q14H), *C158T, C335T (H78Y), *T350G (L83V) | 30 |
| E6 | A169C, G267A, T286A, A289G, G293A, G507A, G525T, A532G | A131G (R10G), *G145T (Q14H), G176A (D25N), T178G (D25E), C315G (S71C), *C335T (H78Y), *T350G (L83V) | 37 |
| E7 | G666A, T789C, T795G, T828C | T732C (F57V) | 27 |
| E7 | G663A, G826T, A746T, T789C, T795G | A647G (N29S), T732C (F57V) | 28 |
| E7 | G663A, A746T, *T789C, *T795G, T843C | A647G (N29S), *T732C (F57V), G823A (G88R), A826T (I89F), T846C (S95L) | 37 |
| LCR | *G7521A, A7636C, C7689A, *T7714G, C7792T, G7826A, A7839C, T7743G, *C7764T, C13T, C7669T, C7678T, A7729C, C7786T, G7799A, G7834T, C7886G | 27 | |
| South India | |||
| L1 | DELETION AND INSERTION AT 6695, 6722,6733, 6736, 6737, 6738 and 6744 | SNPs at 6722, 6742, 6743, 6759, *6760, *6726, *6729, *6730, *6732, *6738, *6739, *6741, *6744, *6759. | 33 |
| L1 | 55595insC, T5597A, G5636A, G5696A, T5801G, A5847C, T5906C, T5909C, T5999G, A6023T, 6028delA, A6068G, A6177C, A6200G, T6245C, 6253delG, 6283insA, G6302A, A6314G, C6539A, T6553C, *C6557T, A6561T, 6564delC, A6581C, A6581T, T6608C, 6658delA, *A6665C, G6719A, G6737T, *C6852T, C6863T, T6911A, T6914C, A6947G, C6968T, *G6992A, A7028G, G7058A, T7076C | *C5562G (Q2E), A5568C (T4P), C5600A (Y14X), A5602G (E15G), A5730C (N58H), *C5862T (H102Y), *C6163A (T202N), A6166T (N2031), G6171A (A205T), A6178C (N207T), C6240G (H228D), A6293C (E245D), C6352T (S265L), A6432G (T292A), A6445C (N296T), A6490C (N311T), C6502T (S315L), A6504C (N316H), T6560A (N334K), 6563delA (K335X), C6564A (P336T), C6565T (P336L), *A6693C (T379P), A6801T (T415S), 6901insAC (448insS), 6950delGT (465delD), A6961C (K468T), C6970A (T471N), A6997G (K480R), *G7058T (L500F), G7084A (G509E), 7093C (K512T) | 37 |
| L1 | 5605delA, 5640delT, G5696A, A5834G, T5909C, T6230C, T6245C, A6314G, A6452G, *C6557T, A6561T, 6654delA, 6663insA, *A6665C, G6719A, *C6852T, C6863T, A6891C, C6968T, T6971G, A6979G, *G6992A, G7058A, 7120insTC | *C5562G (Q2E), T5597G (C13W), T5598G (Y14D), G5607A (D17N), *C5862T (H102Y), *C6163A (T202N), G6171A (A205T), A6178C (N207T), C6240G (H228D), A6432G (T292A), A6458G (D300E), T6560A (N334K), 6563delA (K335X), *A6693C (T379P), A6801T (T415S), 6901insAC (448insS), 6950delGT (465delD), G7008C (D484H), C7011A (L4851), *G7058T (L500F), T7110A (S518T) | 37 |
| L1 | T5673C, A5687G, G5696A, A5702C, A5834G, T5909C, 6022delC, A6023T/C, G6059A, A6068G, 6087delT, T6245C, T6269C, A6314G, T6317A, A6389G, A6452G, T6482C, C6539A, A6554C, *C6557T, A6581T, A6656G, *A6665C, A6668G, G6719A, C6726T, G6836A, *C6852T, T6860C, C6863T, G6893A, A6938C, A6947G, C6968T, A6989G, *G6992A, G7058A, T7076C, T7130A, C7143A, T7145G | C5562G (Q2E), A5620G (Y21C), C5862T (H102Y), *C6163A (T202N), A6178C (N207T), C6240G (H228D), A6432G (T292A), 6563delA (K335X), *A6693C (T379P), A6801T (T415S), T6822C (S422P), 6901insAC (448insS), A6947C (E463D), 6950delGT (465delD), C6957T (L467F), *G7058T (L500F), C7117A (T520N), A7147C (K530T) | 37 |
| E2 | A2983G, A3538C, T3664A, C3684A, T3694A, T3706C, G3778T | C3161T (H136Y), C3516A (T254N), T3517C (T254N), T3566G (F271V), T3371C(S206P), C3159A (T135K), A3362G (N203D), C3410T (P219S), G3449A, G3778A | 32 |
| E6 | 350T, *T350G (L83V), *C335T (H78Y), *G145T (Q14D), T419G (C106G) | 31 | |
| E6 | T286A, A289G, A508C, A532G, A536DEL | G145T (Q14H), T178G (D25E), T329C (Y76H), *C335T (H78Y), G491A (G130S), C528T (S142L) | 37 |
| E6 | T254DEL, T265DEL, T279DEL, T286A, A289G, A532G | G145T (Q14H), T178G (D25E), T179G (I26V), G188A (E29K), *C335T (H78Y), *T350G (L83V), A442C (E113D), A526T (R141S), T527A (S142T) | 37 |
| E6 | T286A, A289G, A532G | G145T (Q14H), T183G (I27R), G205T (K34N), C245T (R48W), T308C (F69L), *C335T (H78Y), *T350G (L83V), T402C (L100S), G491A (G130S) | 37 |
| E7 | T732C, T795G | *A647G (N29S), *C790T (R76C) | 31 |
| E7 | *T789C, *T795G, T843C | A647G (N29S), *T732C (F57V), C747DEL (Frame shift), G842DEL (Frame shift), T844DEL (Frame shift), T846C (S95L) | 37 |
| E7 | INS668GGA, *T789C, *T795G, T843C | A645C (L28F), A647G (N29S), *T732C (F57V), T846C (S95L) | 37 |
| E7 | G585A, T756C, *T789C, *T795G, C854DEL | A619T (T20S), G709A (A50T), C712T (H51Y), *T732C (F57V), C747DEL (Frame shift), A826DEL (Frame shift), A826T (I89F), T844DEL (Frame shift), A852DEL (Frame shift, K97N, P98H) | 37 |
| Central India | |||
| E6 | A229G | GAT114TAC (D4Y), A225G (E41G), C240G (A46G), T244G (F47V), *C315G (S71C), A334T (R77S), *T350G (L83V), T400G (L99V), C422A (Q107K), C523A (C140Stop), G545A (E148K) | 34 |
| E7 | C834T, C836A | T573G (D4E), *G823C (G88R), *T838C (I93T), *T841A (C94S) | 34 |
| East, Central and North India | |||
| L1 | T5675C, *G5698A, A5752C, A5755C, C5864T, *T6247C, A6316G, T6319C, C6370T, A6391G, A6425C, C6541A, C6559T, A6565G, T6661C, A6667C, G6721A, C6826T, *C6854T, C6865T, T6889G, A6952G, C6970T, G6994A, T7027G, G7031T, G7032A, G7060A, A7072T, T7078C, T7123C, A7141C | G6069C (R144T), *C6165A (T176N), G6173A (A179T), A6180C (N181T), C6182G (P182A), *A6434G (T266A), T6438G (V267G), A6445C (E269D), G6458A (D274N), A6492T (N285I), G6582A (R315Q), T6689G (S351A), *A6695C (T353P), A6803T (T389S), T7028G (L464V), *G7060T (L474F), A7148C (K504Q) | 36 |
| L2 | A4242G, A4266C, T4281C, A4299C, A4410G, A4413C, G4428T, T4446G, T4452C, G4461A, A4467C, A4468C, A4504C, C4525T, T4527C, T4545G, G4563A/T, T4572G, A4599C, A4626T, A4632C, T4644A, T4647G, T4695C, *C4725T, A4788C, C4848T, A4887G, G4938A, A4944G, A4950G, A5034T, A5073T, G5142A, A5187C, T5286A, G5379A, T5403C/G, A5409G, T5412G, T5475A, T5478C, C5487T, A5517G, T5523C, A5532G, A5622G | C4253T (S6F), A4368C (Q44H), T4600C (S122P), G4621A (D129N), G4654A (D140N), A4693C (T153P), A4811C (N192T), A4820G (E195S), A4821C (I196L), C4825T & C4826A (P197Y), A4969G (T245A), G5008A (E258K), A5027C (N264T), T5041C (S269P), C5042A (S269Y), A5048C (N271H), A5059G (I275V), T5060A (I275N), A5063C (N276T), A5152C (I306L), *A5226T/C (L330F/F), C5231G (T332S), G5236C/A (D334H/N), A5250C (E338D), A5256T (E340D), A5356C (I374L), T5357G (I374S), T5368G & C5369T (S378V), T5386G (S384A), G5389A (V385I), A5466C (L410F), A5492C (I418T), T5495C (I420T), A5505C (Q423H), G5506A (A424T), C5509T (P425S), A5518C (I428L), C5564G (A443G), G5609A (R458Q) | 36 |
| L2 UTR/NCR2 | A4150C, C4151A, *T4152G/A, A4157G, T4158A, *C4164T, A4184T, T4186A, T4222C, *T4228C/G/A, T4231G, A4233C, A4234C | 36 | |
| E1 | T921C, T1221C, T1446C, G1941A, T2301C, T2343C, C2344T, T2470C, T2478C, T2778C | C874T (P4S), G1293T (Q143H), T1297A (L145I), G1363A (G167S), T1421C (I186T), A1842G (I326M), T1920G/C (D352E/D), G2337A (M491I), G2650A (E596K) | 39 |
| E2 |
T2914C, C3007T, G3058A, A3187C, A3253G, T3259C, T3313G, C3436A, A3538C, T3640C, T3664C, *C3684A, T3694A, T3706C, G3778T/C, C3787A, C3800G, T3805G, T3820C, T3847C |
A2919C (N55T), C2988T (T78M), C3159A (T135K), C3161T (H136Y), T3172G (I139M), A3181C (E142D), G3182A (A143T), A3208C (Q151H), *T3224A (L157I), C3236G (H161D), G3249A (R165Q), A3285C (K177T), C3348T (S198F), A3362G (N203D), A3366C (E204A), T3371C (S206P), C3372A (S206Y), C3377G (P208A), T3387C (I211T), *C3410T (P219S), G3413A (A220T), C3516A (T254N), T3517C (T254N), T3566G (F271V), C3576T (S274L), G3585C (G277A), A3605G (N284D) | 36 |
| E4 | A3362G, T3371C, C3377G, T3387C, *C3410T, G3413A, A3605G | A3366C (K9Q), C3372A (P11T), C3436A (S32stop), C3516A (L59T), T3517C (L59T), A3538C (Q66P), T3566G (H75Q), C3576T (H79Y), G3585C (D82H) | 36 |
| LCR | A7168G, T7176G, T7187C, G7193T, A7197C, A7227C, *A7233C, A7266G/C, A7376T, C7394T, C7395T, T7401C, G7427A, G7435A, *T7450C, A7482G, A7483C, *A7485C, *G7489A, *G7521A, A7550G, C7577T, C7655A, *C7669T, *C7689A, T7713G, T7714G/A, A7729C, T7743G, C7764T, C7792T, A7773C, T7775A, C7786T, G7826A, T7827A, G7834T, G7868A, C7886G, C13T, C24T, A93T, A98C | 36 | |
| West and South India | |||
| L1 |
*C5990T, *C5695T |
A4941C (N56T), C5000T (H76Y), A5049C (N92T), A5248C (L158F), T5307G (V178G), A5316C/T (N181T/I), A5481C (K236T), A6101C (K443Q), A6135C (K454T), A6198G (K475R), C5301A (T176N), G5309A (A179T), *A5570G (T266A), A5628C (N285T), T5662A (S296R), *A5831C (T353P), A5939T (T389S), T5960C (S396P) and *G6196A/T (L474F) | 40 |
| L2 | (L75F), (T85A), (T94A), (S122P/A), (S134R), (T245A), (L266F), *(S269P), (S270N), (D272N), (N273S), (I306L), *(L330F), (T332S), *(D334N), (D334H), (D334T), (E338D), (Q342L), (T352P), (H354Q), (T377S), (S378V/F), (S384A), (V385I), (L390F), (I418M), (I420T), (Q423H), (A424T), (S426A), (I428L), (A443G) | 42 | |
| LCR | *G6657A, *T6586C, *A6865C, A6475T, T6554C, G6606T, C6701T, A6737T, A6771T, C6805G, T6836G, T6847G, A6866C, T6917C, T6926A, T6967A, A6973C, A6975G, G6978A, G6988A, G6992A, G7004C, G7005A, C7012A, T7015C, C7025T, C7055T, C7066T, C6530T, C6531T, A6618G, A6621C, G6625A, T6704G, A6772C, C6805T/G, C6825A, T6849G, T6850G, T6879G, C6900T, C6922T, C6928T, G6962A, G6970T, G7004A, C7022G | 41 | |
| South and East India | |||
| E6 | A532G, T286A, A289G, T421G, G522C | *T350G (L83V), C335T (H78Y), A276C (N58S) | 35 |
| Gene (no. of participants) | Variant lineages and sublineages | Percentage prevalence; % (sample type) | References |
|---|---|---|---|
| East India | |||
| E2 (49 cases and 23 controls) | European (A1-3) |
87.76 (cases) 96.3(controls) |
23 |
|
Asian American (D2-3) |
12.24 (cases) 3.7 (controls) |
||
| LCR (42 cases and 19 controls) | European (A1-3) |
88.09 (cases) 94.73 (controls) |
|
| Asian American (D2-3) |
11.9 (cases) 5.26 (controls) |
||
| Whole genome (145 cases and 24 ontrols) | European (A1) |
86.89 (cases) 100 (controls) |
24 |
| North American (D1) | 4.13 (cases) | ||
| Asian American (D2) | 8.96 (cases) | ||
| West India | |||
| E6 (32 single infections; 20 coinfection and 10 multiple types infections) | European (A1-3) | 90.3 | 25 |
| North American (D1) | 4.8 | ||
| Novel variants | 4.8 | ||
| North India | |||
| L1 (31 cervical cancer patients) | European (A1-3) | 70.9 | 27 |
| Asian American (D2-3) | 12.9 | ||
| North American (D1) | 6.5 | ||
| Novel variants | 9.7 | ||
| E6 (60 cervical cancer patients) | European (A1-3) | 85 | |
| North American (D1) | 10 | ||
| Asian American (D2-3) | 3.3 | ||
| New variants | 1.7 | ||
| E7 (60 cervical cancer patients) | European (A1-3) | 86.7 | |
| African-2 (C) | 11.6 | ||
| Asian (A4) | 1.7 | ||
| LCR (60 cervical cancer patients) | European (A1-3) | 63.3 | |
| Asian American (D2-3) | 13.3 | ||
| African-1 (B) | 11.6 | ||
| African-2 (C) | 3.3 | ||
| Novel variants | 8.3 | ||
| E6 (74 cervical cancer; 9 SILs; 9 controls) | European (A1-3) |
86.4 (cases) 100 (SIL) 88.9 (controls) |
28 |
| Asian American (D2-3) |
10.8 (cases) 11.11 (controls) |
||
| Asian (A4) | 1.35 (cases) | ||
| African-2 (C) | 1.35 (cases) | ||
| E6 (221 cervical cancer cases) | European (A1-3) | 97.25 | 30 |
| Asian (A4) | 1.5 | ||
| Asian American (D2-3) | 0.75 | ||
| South India | |||
| L1 (57 samples) | European (A1-3) | 48 | 33 |
| CENTRAL INDIA | |||
| E6, E7 (21 cervical cancer cases) | European (A1-3) | 52.38 | 34 |
| Asian (A4) | 47.61 | ||
| Multiregional | |||
| E6 (50 cases and 20 controls) | European (A1-3) | 92% (cases) & 100% (controls) | 35 |
| North American (D1) | 4 | ||
| Asian American (D2-3) | 4 | ||
|
E6, E7, and L1 (412 cervical cancer patients; Delhi-84 Mumbai-79 Kolkata-54 Bangalore-69 Thiruvananthapuram-59 Vellore-67) |
European (A1-3) |
86.8 (Delhi- 86.9 Mumbai-83.54 Kolkata-85.18 Bangalore-89.85 Thiruvananthapuram-81.35 Vellore-94.02) |
37 |
| Asian American (D2-3) |
11.4 (Delhi-10.7 Mumbai-16.4 Kolkata-14.8 Bangalore-7.2 Thiruvananthapuram-13.6 Vellore-6) |
||
| Asian (A4) |
1.7 (Delhi-2.38 Mumbai-0 Kolkata-0 Bangalore-2.9 Thiruvananthapuram-5.1 Vellore-0) |
||
| E1 (70 samples) | European (A1-3) | 81.4 | 39 |
| Asian American (D2-3) | 18.6 | ||
| L1 (133 samples) | A | 86.96 | 40 |
| European (A1) | 48.1 | ||
| European (A2) | 26.3 | ||
| European (A3) | 0.75 | ||
| Asian (A4) | 9.8 | ||
| D | 15.03 | ||
| North American1 (D1) | 1.5 | ||
| Asian American1 (D3) | 13.5 | ||
|
LCR (as per L1 gene) (84 isolates) |
European and/or Asian (A) | 84.5 | 41 |
| North American and/or Asian American (D) | 15.5 | ||
|
L2 (as per L1) (124 isolates) |
European and/or Asian (A) | 86.3 | 42 |
| North American and/or Asian American (D) | 13.7 | ||
| Region | Nucleotide variations | Lineage | Ref | |
|---|---|---|---|---|
| Synonymous variations | Non-synonymous variations | |||
| E6/E7 gene variations | ||||
| North India | A89C, A92G, T104C, T116C, A218T, *T485C, A536C, *C549A, C751T | T553G, T858G | European (A1-3) | 28 |
HPV 16 variants
Variants in East India
A study reported 20 variations within the E2 gene with C3007T, G3058A, C3236G, A3253G, and A3605Gas prevalent among controls, depicting that these may confer reduced pathogenicity in HPV16 variants23. Certain novel variations were also observed. Additionally, specific E2 variations were used by the authors to determine lineages, which suggested that the majority of the variants belonged to the European (A1-3) lineage followed by the Asian American lineage (D2-3). It was also reported that E2 disruption was more frequent among cases. In addition, 22 variations in the long control region (LCR) region were also determined. Variation T7450C was reported to be present in E2 binding site-IV and was prevalent among cases. In addition, most variants having this variation had no other variation within the LCR or E2 gene, thus indicating inter-gene co-variations within individual isolates. The majority of LCR variants were reported to belong to European (A1-3) lineage, followed by Asian American (D2-3).
Another study from eastern India observed that 262 single nucleotide variants were present within the studied samples, among which 20 non-synonymous variations were deleterious24. The most prevalent variations are depicted in table III (with MAF ≥0.05; the variations whose synonymous/non-synonymous status could not be deduced were omitted from the table). The lineages and sub-lineages were assigned as per E2 (T3694A), E6 (A532G) and LCR (T7743G, G7834T) variants. The most prevalent lineage among the samples was European (A1), followed by North American (D1) and Asian American (D2). The D1 and D2 sub-lineages were reported to be exclusively present within the CC cases. Furthermore, it was reported that lineages A and D could be distinguished by 35 bi-allelic variants, while 6 variants could distinguish D1 and D2 sub-lineages. Subsequently, it was observed that certain variants from the A1 sub-lineage had differential distribution among malignant and non-malignant samples and could be associated with pathogenicity. To further understand the viral genomes in the Indian population, network analysis was performed, and it was found that four haplotypes were present among the A1 sub-lineage, among which two haplotypes were specific to cases. Additionally, certain nucleotide variations were co-existing. The propensity of D lineage towards integration of the HPV genome was suggested. Consequently, the presence of E5 variants (3979C, A4042G) and LCR region variations (7577T) in conjunction with E6 350G variations as risk alleles suggested the polygenic aspect of viral contribution to disease.
Variants in West India
A study reported the prevalence of the European T350G variant in participants having coinfections with other HPV types, while the North American variant was observed only in women with coinfections with HPV16 and 33, suggesting that these variants may be facilitating coinfections with other HPV types25. In addition, 3 novel variations were observed. Phylogenetically, it was reported that the European T350G variant was the most predominant variant, followed by the European prototype (A1-3), North American (D1), and novel variants, based on the classification given by Yamada et al26.
Variants in North India
A study analyzed the partial L1 and entire E6, E7, and LCR sequence variations among participants27. Of 13 L1 variations, the most prevalent variation was A6695C (54.5%). Four novel variants were also observed. Furthermore, novel L1 variants with A6667C and A6691G variations did not vary within E6, E7, and LCR regions. The authors classified identified variants based on the classification given by Yamada et al26. Most of the L1 variants belonged to the European (A1-3) lineage. Furthermore, 42 patients showed variations in the E6 gene. T350G variant was predominantly found in all the cases, either alone (81%) or with other variations (19%). Additional variations were observed only in variants consisting of T350G variation. Most of the E6 variants belonged to the European (A1-3) lineage. Additionally, five E7 variations were found only in eight cases, with the majority belonging to the European (A1-3) lineage. Furthermore, 45 cases comprised variations within the LCR region. The most frequent variation was G7521A (91.1%), followed by C7764T (22.2%) and T7714G (20%), mostly found along with G7521A. Four novel variants were observed in participants. Predominant lineages as per LCR were European (A1-3), followed by Asian American (D2-3), African-1(B), and African-2 (C).
Another study determined variations within E6 and E7 genes28. Fourteen variations within the E6 gene were observed, of which four were novel in the Indian subcontinent and were observed exclusively among cases, indicating that these may be associated with oncogenicity. In addition, seven variations were observed within the E7 gene, of which two were novel. The majority of the variants, as per the classification29, were reported to belong to European (A1-3) lineage followed by Asian American (D2-3).
Additionally, another study reported six variations in the E6 gene30. T350G (67.5%) variation was predominant, followed by C158T (15%) and G145T, T286A, and C335T (12.5%, each). As stated by the authors, European (A1-3) variants were found to be predominant.
Variants in South India
A study reported variations within E6 and E7 genes31. Six variants of the E6 gene were identified. Two variants had European prototype gene sequences, i.e., 350T (9.1%) and T350G variant (19.6%), respectively. Furthermore, another variant (14.7%) had two variations, i.e., T350G and C335T. The fourth variant (19.6%) had variations at T350G along with G145T. The fifth variant (28.7%) had variations, T350G, C335T, and G145T. The sixth variant (8.4%) observed had a variation of T419G as well as T350G. Additionally, some variants were found to be more predominant in CC patients than in controls, suggesting their association with the disease. It was noted that co-variations play an important role in determining the pathogenicity of a variant. Furthermore, one variant was prevalent in younger age group patients, i.e., below 45 yr, pointing towards its aggressive nature. In addition, four variants of E7 were found in this study. One variant had variation at A647G (37.8%). Other variants had variations T732C (21.7%), C790T (23%), and T795G (17.5%), respectively.
Another study stated that 38 samples with intact E2 genes were used for variant analysis32. Variants were divided into four groups based on cleavage by different restriction endonucleases. Sequencing of four samples from each restriction fragment length polymorphism (RFLP) group showed that four isolates from group one did not have any variation, while 10 isolates had variation A2983G. The third group comprised 11 samples with 13 variations. Thirteen samples from the fourth group comprised 12 variations.
Similarly, the distribution of HPV types was determined among cervical samples from non-malignant, pre-malignant (LSIL and HSIL) and malignant samples (squamous and adenocarcinoma)33. Sequence variations were analyzed among HPV16-positive samples within the L1 gene. Nucleotide variations involved deletions and insertions as well as single nucleotide polymorphisms (SNPs). Some variations at 6726, 6729, 6730, 6732, 6738, 6739, 6741, 6742, 6744, 6759, and 6760 were found to be predominant among patients with single and multiple HPV infections. Phylogenetic analysis of 57 HPV16 positive samples along with 12 reference sequences led to the clustering of samples into three groups comprising 48.5, 29, and 18.9 per cent of samples, respectively. In addition, as per alignment with reference sequences with known lineages, 48 per cent of samples were reported to be aligned to the European (A1-3) lineage.
Variants in Central India
A study reported 12 variations within the E6 gene34. The most prevalent variation was T350G (76.2%), followed by C315G (28.6%). In addition, six variations within the E7 gene of eight HPV16 isolates were observed. The most common variation was G823C (23.8%), followed by C838A and T841A (9.5%, each). The variants were stated to belong to European (A1-3) and Asian (A4) lineage.
Variants in multiple regions in India
A study observed eight variations within the E6 gene among 38 patients35. T350G was the predominant variant in cervical neoplasia compared to controls. Three novel variations were reported. Only three controls harboured variations within the E6 gene. The variants, as per the classification26, were reported to belong to European (A1-3), Asian American (D2-3) and North American (D1) lineage.
Another group stated 49 variations within the L1 gene of HPV1636. The most common variations observed were G7060T with minor allele frequency (MAF) of 0.84, followed by A6434G and C6854T (MAF=0.16, each) and G5698A, C6165A, T6247C, A6695C, (MAF=0.14, each). In addition, there were 92 variations within the L2 gene. Non-synonymous variations in the L2 region were higher among CC cases. Common variations within the coding region were A5226T (MAF=0.64), A5226C (MAF=0.29), and C4725T (MAF=0.16). Furthermore, 48 variations were observed within E2 and E4 genes. The most common variations observed were C3410T (MAF=0.17), C3684A (MAF=0.16) and T3224A (MAF=0.14). In addition, 45 variations within LCR were observed. Predominant variations observed were T7450C (MAF=0.45), followed by G7521A (MAF=0.16) and A7233C, A7485C, G7489A, C7669T and C7689A (MAF=0.13, each). Additionally, 13 variations within L2-UTR or NCR2 were reported, with certain common variations such as T4152G (MAF=0.97), T4228C (MAF=0.76), and C4164T (MAF=0.15).
A multicentric study reported 204 variations within the L1 gene of HPV1637. Common non-synonymous variations were C6163A (13.9%), A6693C (12.4%), C5562G (12.2%), C5862T (12.2%), A6178C (9.7%) and G7058T (11.3%). However, common synonymous variations were C6852T (14.3%), A6665C (13.8%), G6992A (13.06%), and C6557T (12.6%). Phylogenetically, European (A1-3), Asian American (D2-3) and Asian (A4) lineages were reported, as per the given classification38. Asian lineage was found only in Delhi, Bangalore, and Thiruvananthapuram samples. In addition, 56 variations were observed within the E6 gene. The most common non-synonymous variations were T350G (72.3%), followed by G145T (13.1%) and C335T (12.1%). Similarly, frequent synonymous variations were T286A (12.1%), A289G (11.7%), and A532G (8.3%). Five novel variations were also observed. Furthermore, 29 E7 gene variations were observed. Commonly observed variations were T789C (11.9%), T795G (11.9%), and T732C (9%). Additionally, six novel variations were observed.
Another study has reported variations within the E1 gene of the HPV16 genome39. Out of 20 variations reported, G1293T (MAF=0.37), T1421C (MAF=0.21) and T921C, A1842G, C2344T (MAF=0.13, each) were commonly observed. Phylogenetic analysis, as per the previous study23, demonstrated that among episomal E2 isolates, European (A1-3) and Asian American (D2-3) lineages were found. In addition, it was reported that variations within the NCR-2 region were higher among E2 intact cases as compared to controls. No microRNA (miRNA) binding site loss was observed among Asian American (D2-3) variants. Furthermore, no mRNA and protein expression of L2 was observed among cases harbouring integrated viral genome.
Another study observed 61 nucleotide variations within the L1 region in patients as well as controls40. The most common variations observed were A5570G (73.3%), G6196A/T (26.3%), C5990T (17.1%), C5695T (15.1%), and A5831C (15.1%). In addition, certain variations were reported for the first time in the Indian population. In addition, L1 variations A5316C/T, A5831C, and A5939T, as well as variants belonging to Asian American1 (D3) lineage, were associated with HSIL. Phylogeny was determined based on the L1 gene, along with certain reference sequences with known lineages, and it was found that 19 L1 sequences were incomplete and, therefore, their phylogeny could not be determined. Out of the remaining samples with complete L1 gene, 113 samples clustered with members of lineage A (European and/or Asian). However, 20 samples clustered with members of D (North American and/or Asian American) lineage. However, it was suggested that the entire genome should be used to ensure the phylogenetic distribution of variants.
Furthermore, another study observed 47 variations within LCR in studied samples, including 23 non-synonymous and 24 synonymous variations41. These variations were present in 64.8 per cent of samples with a normal cervix, 88 per cent of women with LSIL, and 89.8 per cent of women with HSIL. The most common variations were G6657A (69.3%), T6586C (27%), and A6865C (18.4%). Certain novel variations were also observed in Indian women. Furthermore, T6586C, G6657A, and T6850G variations were associated with HSIL. In addition, multiple variations were common in LCR from women having HSIL. Phylogenetically, the Asian (A4) sub-lineage was reported to have multiple variations related to HSIL. The phylogeny of variants was determined based on the L1 gene and by using reference sequences with known lineages. The corresponding L1 sequence of 18 isolates was not available, and thus, lineage could not be assigned. Among the remaining isolates, 84 clustered as per assigned lineages.
Another study observed 91 variations within the L2 gene, of which 43 were non-synonymous and 48 were synonymous42. These variations further translated to 35 non-synonymous substitutions and 53 synonymous substitutions. Substitutions L330F (75.6%), S269P (28.4%), and D334N (24.3%) were commonly observed. Sixteen substitutions reported in Indian isolates were novel. It was also reported that samples with multiple substitutions were associated with HSIL. Additionally, substitutions S384A, L266F, S378V, and T245A were also associated with HSIL and were further reported to be predominant in Asian American (D3) sub-lineage, substantiating its aggressive nature. Furthermore, substitutions T245A, L266F, S378V, and S384A were reported co-mutating in 19 samples. In this study, lineages were assigned to 134 of the total samples based on the L1 gene; 14 isolates without corresponding L1 gene were not assigned lineage. These 14 isolates, however, clustered with lineages A (European and/or Asian) and D (North American and/or Asian American) equally. Of the isolates with assigned lineage, 124 clustered as per assigned lineage. However, there was no difference in the predominance of non-synonymous variations between normal and HSIL cervical statuses, as reported by Bhattacharjee et al36.
HPV 18 variants
A study28 from North India reported variations within the E6 and E7 genes of HPV 18. The study stated that all of these variants belonged to the European (A1-3) class, as per the classification by Ong et al43. Common E6 and E7 variations observed were T485C and C549A among 13 samples.
Putative functional significance of HPV variants
Nine non-synonymous variations in L1 protein were observed in immune-dominant loops within L1 protein40. Many of the variations were predicted to affect B-cell and /or T-cell binding, impairing the human leukocyte antigen (HLA)-interaction, while others were impacting the stability of 3D protein structure as well as L1 monomer and pentamer (Table VI)37,40. There were 29 deleterious non-synonymous variations within the HPV16 genome, among which 26 were exclusively present within CC cases, while three were also present in controls36. Among these deleterious variations, 17 were present within L1, L2, and E2 genes. Variations impacting B/T- -cell epitopes were also observed in L2 gene42.
| Gene | Substitutions | Location/Function |
|---|---|---|
| L1 | N56T, N92T, L158F, T176N, V178G, A179T, N181T/I, N285T, S396P, K454T | B-cell epitope |
| L1 | H76Y, T266A, N285T, S296R | T-cell epitope |
| L1 | L158F, A179T, K236T, T266A, S296R, T353P, S396P, K454T | Destabilize the 3D structure of the protein |
| L1 | N56T, H76Y, N92T, T176N, V178G, N181T/I, N285T, T389S, K443Q, L474F | Stabilize L1 monomer |
| L1 | T266A, T353P | Affects L1 pentamer stability |
| L2 | L75F, T85A, T94A, T254A | B-cell epitope |
| L2 | T85A, S122P/A, S134R, T245A, L266F, S269P, S270N, I306L | T-cell epitope |
| E6 | F69L, M137L | B-cell epitope |
| E6 | H78Y, L83V |
Affect epitopes of MHC I & II Creates additional HLA epitopes |
| E6 |
L83V+F69L L83V+M137L |
Destabilizes E6AP complex |
| LCR | G7521A, C7786T | YY1 and YY14 binding site |
| LCR | T7714G, T7743G, C7764T | NF1 binding site |
| LCR | G7004A/C, G7868A | E2 binding site |
MHC, major histocompatibility complex; HLA, human leukocyte antigen; E6AP, E6 associated protein; LCR, long control region
Similarly, the variations observed in the E6 protein were reported to affect B-cell epitopes, stability of the E6AP complex, and binding affinity of peptides to Major histocompatibility complex (MHC). In contrast, some variations were reported to create additional epitopes for HLA5,30. The amino acid variations within the epitope regions may have altered immune response, and thus, the vaccines designed for a particular variant may have reduced efficacy against the variants found in other regions. Furthermore, 3D modelling predicted that variants with C523A variation within the E6 lack the last 12 amino acids of the second domain, which made the hook-like structure and interact with amino residues of the preceding domain34. Thus, further studies must be conducted to evaluate the epitopes that can be affected by such variations for the rational design of HPV vaccines.
The significance of LCR variations was determined by their localization in various Transcription factor (TF) binding sites. Several variations were found in known binding sites of various TFs23,26,41. These variations in the binding sites of various TFs were also reported to be associated with particular lineages. In addition, it was also reported that NCR-2 of HPV16 consists of 14 binding sites for miRNA39. European variants, among those having intact genome, were reported to have T4228C variation, which was linked with the loss of nine miRNA binding sites.
Discussion
The viral early genes are associated with transformation and oncogenicity, while the late genes are related to capsid formation and the non-coding regions perform regulatory function6,36,44. Hence, any variation within HPV genes may affect its transformational potential.
Studies have reported that different variants of HPV have varying oncogenic potential and risk associations. For instance, non-European variants of HPV16 are associated with an increased risk of persistence, cervical intraepithelial neoplasia (CIN), and cancer development13. In addition, these variants have also been reported to be associated with histologic subtypes.
Upon analyzing the results of the phylogeny of variants from the given studies, it was found that the distribution of lineages varies with the different genomic regions sequenced. However, all such studies have relied on sequencing specific genes of the viral genomes, such as L1, E2, E6, E5, LCR, etc., either singly or in combinations of two or more genes, rather than whole viral genome sequencing. Thus, it is required to sequence the complete genome to ascertain the specific lineage of the variants. Additionally, the risk of variants varies with the ethnicity of the infected population, as Asian women infected with the A4 sublineage and Hispanic women with D2/3 sublineages were reported to have a higher risk of CIN3+ compared to other ethnic groups45. Although nucleotide variations within a particular gene might be associated with disease susceptibility, a haplotype comprising such variation might not depict significant association, thus posing difficulty in ascertaining risk association and selecting biomarkers36.
A critical analysis of all these studies indicated that there are certain unique as well as common variations in the HPV genome with respect to geographical regions (Fig. 2A-C). Here, the studies in which there was no segregation of patients from different regions, as well as those genes for which less than four geographical regions were represented, were excluded from the analysis. Furthermore, it can be noted that there are more exclusive variations in southern India as compared to other regions, which could be attributed to the sample size as well as the inclusion of more than one city in the southern region. Thus, suggesting that more studies need to be done in this direction from other regions of India.
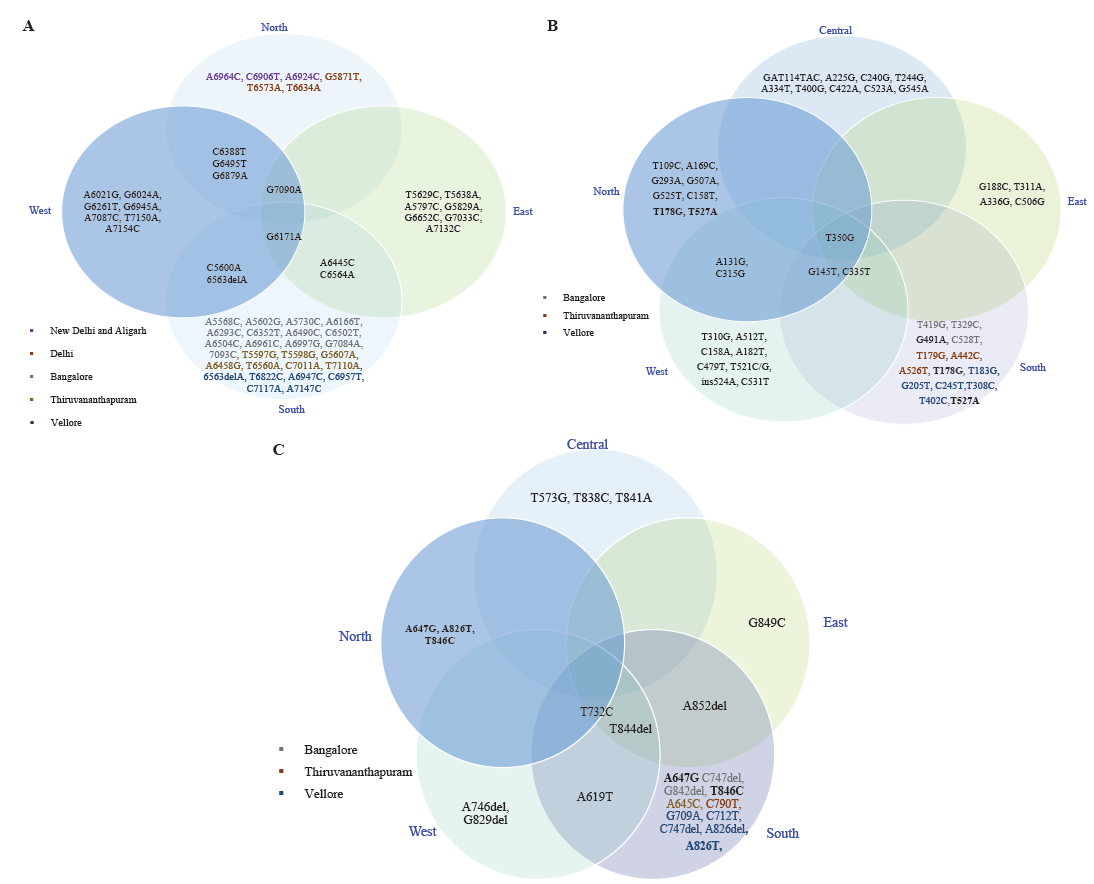
- Distribution of HPV16 non-synonymous variations within different regions of India (A) L1 variations, (B) E6 variations, and (C) E7 variations (different colors in text represents different locations from the same region).
In addition, there were certain variations within the HPV16 genomes that were common in different regions (Fig. 3A-C); however, their prevalence varies among them. For instance, C3161T, T3517C, T3566G, and T3694A variations in the E2 region were relatively more prevalent in south India29, and C3684A was more frequent in east India23. The differences in variation patterns observed in various genomic regions may be attributed to regional characteristics, ethnicity of the population as well as geographical conditions. Keeping in view HPV variants and their antigenicity, it has been reported that amino acid substitutions within certain HPV variants may affect the immune response and thus, vaccines developed against one variant may have reduced efficacy in regions where these variants are less prevalent7. Furthermore, a study was designed to assess cross-protection offered by HPV Bivalent/Quadrivalent vaccine towards variants of genetically related hr-HPV infections16,45. It was demonstrated that the vaccine-induced antibodies depicted reduced neutralization of HPV 45 (B2 variants)16. Similarly, the vaccine efficacy against HPV 31 lineage A/B was partial for transient infections and not efficacious for persistent infections45. While, in other studies, altered neutralization against HPV 33 (A2, A3, B, and C variants), HPV 52 (D variants), and HPV 58 (C variants) was observed17-19. This variability in neutralization was observed to occur because of specific residues located within the immune dominant loops of the L1 protein. In contrast to the extensive studies available for some other viruses46,47, limited evidence is available regarding vaccine escape of the HPV variants. Therefore, additional studies should be carried out to evaluate the impact of HPV genomic variations within the epitope region of various genes for rational vaccine design. In addition, the biological significance of the variants with such variations needs to be validated through experimental studies.
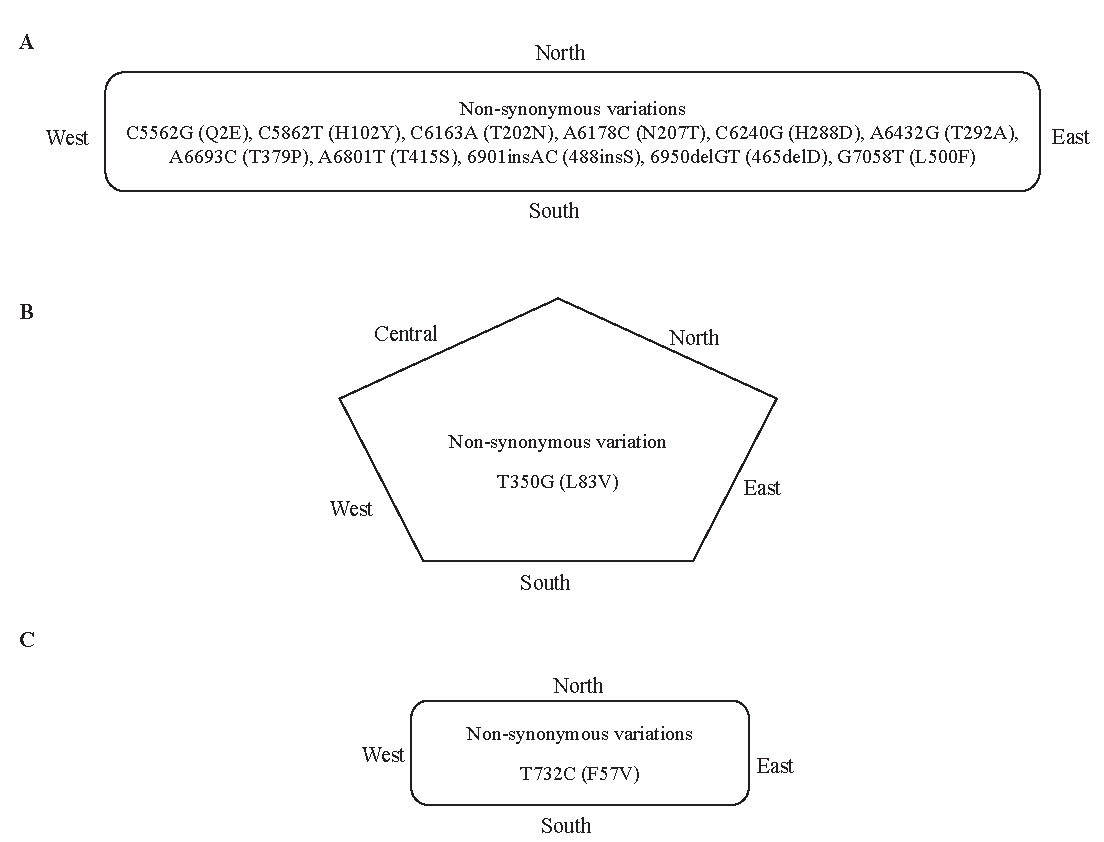
- Common variations found in different regions of India (A) L1 variations, (B) E6 variations, and (C) E7 variations.
Conclusion
The present compilation highlights some significant findings, including regional variations in HPV variants and their putative functional significance. Comprehensive studies encompassing participants from different regions and ethnicities in a large data set are required to ascertain clinically significant HPV variants. The majority of studies reported hitherto have focused on particular genes for variant identification rather than the entire viral genome. Therefore, further studies involving the whole viral genome approach are required to ascertain the distribution of variants from different geographical regions as well as for better identification of polygenic aspects of viral contribution to diseases. In addition to HPV-16 and -18, studies also indicate the involvement of other hr-HPV types, so while planning such studies involving variants detection, these HPV types should also be taken into consideration. This will help not only in better diagnosis and management but also in vaccine design for HPV. Keeping in consideration that HPV is not only a menace for CC but many other oncogenic conditions, such studies should also be carried out in other relevant disease conditions.
Acknowledgment
First author (NS) acknowledges the Council of Scientific & Industrial Research for providing fellowship to pursue PhD research.
Financial support & sponsorship
None.
Conflicts of Interest
None.
Use of Artificial Intelligence (AI)-Assisted Technology for manuscript preparation
The authors confirm that there was no use of AI-assisted technology for assisting in the writing of the manuscript and no images were manipulated using AI.
References
- Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71:209-49.
- [CrossRef] [PubMed] [Google Scholar]
- Global Cancer Observatory: Cancer Today. Available from: https://gco.iarc.who.int/today, accessed on February 1, 2024.
- Epidemiology of human papillomavirus related cancers in India: Findings from the National Cancer Registry Programme. Ecancermedicalscience. 2022;16:1444.
- [CrossRef] [PubMed] [PubMed Central] [Google Scholar]
- Human papillomavirus molecular biology and disease association. Rev Med Virol. 2015;25:2-3.
- [CrossRef] [PubMed] [PubMed Central] [Google Scholar]
- HPV16 sublineage associations with histology-specific cancer risk using HPV whole-genome sequences in 3200 women. JNCI: J Natl Cancer Inst. 2016;108:djw100.
- [CrossRef] [Google Scholar]
- Mechanistic role of HPV-associated early proteins in cervical cancer: Molecular pathways and targeted therapeutic strategies. Crit Rev Oncol Hematol. 2022;174:103675.
- [CrossRef] [PubMed] [Google Scholar]
- Epidemiology and burden of human papillomavirus and related diseases, molecular pathogenesis, and vaccine evaluation. Front Public Health. 2021;8:552028.
- [CrossRef] [PubMed] [PubMed Central] [Google Scholar]
- Epidemiological and functional implications of molecular variants of human papillomavirus. Braz J Med Biol Res. 2006;39:707-17.
- [CrossRef] [PubMed] [Google Scholar]
- Vaccine escape challenges virus prevention: The example of two vaccine‐preventable oncogenic viruses. J Med Virol. 2023;95:e29184.
- [CrossRef] [PubMed] [Google Scholar]
- The genetic drift of human papillomavirus type 16 is a means of reconstructing prehistoric viral spread and the movement of ancient human populations. J Virol. 1993;67:6413-23.
- [CrossRef] [PubMed] [Google Scholar]
- The clinical importance of the nomenclature, evolution and taxonomy of human papillomaviruses. J Clin Virol. 2005;32:1-6.
- [CrossRef] [PubMed] [PubMed Central] [Google Scholar]
- Mechanisms of human papillomavirus-induced oncogenesis. J Virol. 2004;78:11451-60.
- [CrossRef] [PubMed] [PubMed Central] [Google Scholar]
- Epidemiology and molecular biology of HPV variants in cervical cancer: The state of the art in Mexico. Int J Mol Sci. 2022;23:8566.
- [CrossRef] [PubMed] [PubMed Central] [Google Scholar]
- Naturally occurring major and minor capsid protein variants of human papillomavirus 45 (HPV45): differential recognition by cross-neutralizing antibodies generated by HPV vaccines. J Virol. 2016;90:3247-52.
- [CrossRef] [Google Scholar]
- Impact of naturally occurring variation in the human papillomavirus 58 capsid proteins on recognition by type-specific neutralizing antibodies. J Infect Dis. 2018;218:1611-21.
- [CrossRef] [PubMed] [Google Scholar]
- Sensitivity of human papillomavirus (HPV) lineage and sublineage variant pseudoviruses to neutralization by nonavalent vaccine antibodies. J Infect Dis. 2019;220:1940-5.
- [CrossRef] [PubMed] [PubMed Central] [Google Scholar]
- Comprehensive assessment of the antigenic impact of human papillomavirus lineage variation on recognition by neutralizing monoclonal antibodies raised against lineage a major capsid proteins of vaccine-related genotypes. J Virol. 2020;94:10-128.
- [CrossRef] [Google Scholar]
- Naturally occurring single amino acid substitution in the L1 major capsid protein of human papillomavirus type 16: Alteration of susceptibility to antibody-mediated neutralization. J Infect Dis. 2017;216:867-76.
- [CrossRef] [PubMed] [Google Scholar]
- The Newcastle-Ottawa Scale (NOS) for assessing the quality of non-randomized studies in meta-analyses. Available from: http://www.ohri.ca/programs/clinical_ epidemiology/oxford.asp, accessed on June 12, 2024.
- Methodological index for non‐randomized studies (minors): Development and validation of a new instrument. ANZJ Surg. 2003;73:712-6.
- [Google Scholar]
- HPV16 E2 gene disruption and polymorphisms of E2 and LCR: Some significant associations with cervical cancer in Indian women. Gynecol Oncol. 2006;100:372-8.
- [CrossRef] [PubMed] [Google Scholar]
- Predominance of genomically defined A lineage of HPV16 over D lineage in Indian patients from eastern India with squamous cell carcinoma of the cervix in association with distinct oncogenic phenotypes. Transl Oncol. 2022;15:101256.
- [CrossRef] [PubMed] [PubMed Central] [Google Scholar]
- HPV16 E6 variants: frequency, association with HPV types and in silico analysis of the identified novel variants. J Med Virol. 2014;86:968-74.
- [CrossRef] [PubMed] [Google Scholar]
- Human papillomavirus type 16 sequence variation in cervical cancers: A worldwide perspective. J Virol. 1997;71:2463-72.
- [CrossRef] [PubMed] [Google Scholar]
- Human papillomavirus type 16 variant analysis of E6, E7, and L1 genes and long control region in biopsy samples from cervical cancer patients in north India. J Clin Microbiol. 2008;46:1060-6.
- [CrossRef] [PubMed] [PubMed Central] [Google Scholar]
- Human papillomavirus genotyping, variants and viral load in tumors, squamous intraepithelial lesions, and controls in a north Indian population subset. Int J Gynecol Cancer. 2009;19:1642-8.
- [CrossRef] [PubMed] [Google Scholar]
- Human papillomavirus type 16 variant lineages in United States populations characterized by nucleotide sequence analysis of the E6, L2, and L1 coding segments. J Virol. 1995;69:7743-53.
- [CrossRef] [PubMed] [Google Scholar]
- Identification of human papillomavirus-16 E6 variation in cervical cancer and their impact on T and B cell epitopes. J Virol Methods. 2015;218:51-8.
- [CrossRef] [PubMed] [Google Scholar]
- Human papillomavirus type 16 E6 and E7 gene variations in Indian cervical cancer. Gynecol Oncol. 2002;87:268-73.
- [CrossRef] [PubMed] [Google Scholar]
- E2 sequence variations of HPV 16 among patients with cervical neoplasia seen in the Indian subcontinent. Gynecol Oncol. 2004;95:363-9.
- [CrossRef] [PubMed] [Google Scholar]
- Prevalence of human papillomavirus types and phylogenetic analysis of HPV-16 L1 variants from Southern India. Asian Pac J Cancer Prev. 2015;16:2073-80.
- [CrossRef] [PubMed] [Google Scholar]
- Genetic analysis of human papilloma virus 16 E6/E7 variants obtained from cervical cancer cases in Chhattisgarh, a central state of India. Virus disease. 2021;32:492-503.
- [CrossRef] [PubMed] [PubMed Central] [Google Scholar]
- HPV 16 E6 sequence variations in Indian patients with cervical neoplasia. Cancer Lett. 2005;229:93-9.
- [CrossRef] [PubMed] [Google Scholar]
- Characterization of sequence variations within HPV16 isolates among Indian women: prediction of causal role of rare non-synonymous variations within intact isolates in cervical cancer pathogenesis. Virology (Lond). 2008;377:143-50.
- [CrossRef] [Google Scholar]
- Molecular variants of HPV-16 associated with cervical cancer in Indian population. Int J Cancer. 2009;125:91-103.
- [CrossRef] [PubMed] [Google Scholar]
- Human papillomavirus type 16 variants and risk of cervical cancer. J Natl Cancer Inst. 2001;93:315-8.
- [CrossRef] [PubMed] [Google Scholar]
- Differential expression of HPV16 L2 gene in cervical cancers harboring episomal HPV16 genomes: influence of synonymous and non-coding region variations. PLoS One. 2013;8:e65647.
- [CrossRef] [PubMed] [PubMed Central] [Google Scholar]
- Characterization of major capsid protein (L1) variants of Human papillomavirus type 16 by cervical neoplastic status in Indian women: Phylogenetic and functional analysis. J Med Virol. 2020;92:1303-8.
- [CrossRef] [PubMed] [Google Scholar]
- Genetic variations in the long control region of human papillomavirus type 16 isolates from India: implications for cervical carcinogenesis. J Med Microbiol. 2022;71:001475.
- [CrossRef] [Google Scholar]
- Genetic variability in minor capsid protein (L2 gene) of human papillomavirus type 16 among Indian women. Med Microbiol Immunol. 2022;211:153-60.
- [CrossRef] [PubMed] [PubMed Central] [Google Scholar]
- Evolution of human papillomavirus type 18: an ancient phylogenetic root in Africa and intratype diversity reflect coevolution with human ethnic groups. J Virol. 1993;67:6424-31.
- [CrossRef] [PubMed] [Google Scholar]
- Key molecular events in cervical cancer development. Medicina (Mex). 2019;55:384.
- [CrossRef] [Google Scholar]
- Cross-protection of the bivalent human papillomavirus (HPV) vaccine against variants of genetically related high-risk HPV infections. J Infect Dis. 2016;213:939-47.
- [CrossRef] [PubMed] [PubMed Central] [Google Scholar]
- Rotaviruses: is their surveillance needed? Vaccine. 2014;32:3367-78.
- [CrossRef] [PubMed] [Google Scholar]
- Vaccine escape challenges virus prevention: The example of two vaccine-preventable oncogenic viruses. J Med Virol. 2023;95:e29184.
- [CrossRef] [PubMed] [Google Scholar]






