Translate this page into:
Molecular characterization of Mumps virus genotype C detected from Dibrugarh district of Assam, India
For correspondence: Dr Prasanta Kumar Borah, Department of Epidemiology and Nutrition, ICMR-Regional Medical Research Centre, NE Region, Dibrugarh 786 001, Assam, India e-mail: prasant47@yahoo.com
-
Received: ,
Abstract
Background & objectives
Mumps, a contagious disease caused by the mumps virus (MuV) involves parotid gland inflammation, with potential complications affecting organs other than the parotid glands and central nervous system. Despite successful vaccination, a resurgence of mumps occurred, raising concerns about vaccine effectiveness. This study aimed to examine the entire genome of a representative MuV genotype C from Dibrugarh, Assam, and compare it with references to detect genetic variations in the circulating strain.
Methods
Representative MuV genotype C from our published study was subjected to whole genome sequencing. MuV genome was analyzed against the reference genome and vaccine strains before being subjected to mutational profiling, N-glycosylation site determination, and phylogenetic analysis. The Immune Epitope Database was used for epitope screening, and selected epitopes were mapped against Assam MuV for conservancy studies.
Results
Mutational analysis of Assam MuV with WHO (World health Organization) reference, vaccine strains Jeryl Lynn (Genotype A), and L Zagreb (Genotype N) showed variations in seven genes. Phylogenetic analysis established Assam MuV as genotype C. Epitope conservancy analysis highlighted subtle variations in experimentally determined T-cell epitopes for HN and F proteins, emphasizing overall epitope stability.
Interpretation & conclusions
Genome sequencing has evolved into a standard and potent method for investigating and recording circulating MuV as it provides information on surveillance, mutation analysis, and transmission dynamics. Despite mumps’ global effect, genomic studies are limited, particularly in north-east. Our study provides first comprehensive whole-genome report on circulating MuV genotype C in Assam. This research contributes vital genomic data, filling gaps in MuV genetic epidemiology, supporting global research, and assessing vaccine effectiveness.
Keywords
Epitopes
genotype C
mumps virus
parotitis
phylogenetic tree
whole genome sequencing
Mumps virus (MuV), a member of the genus Rubulavirus in the Paramyxoviridae family is responsible for causing highly contagious disease called m umps. The MuV genome is composed of 15,384 nucleotides and is non-segmented, single-stranded, negative-sense enveloped RNA. Seven transcription units are linked in tandem makeup of the MuV protein: nucleocapsid (N), phosphorylated (P), matrix (M), fusion (F), small hydrophobic (SH), hemagglutinin-neuraminidase (HN), and large (L)1. Among these seven genes, SH gene exhibits greatest variability in nucleotide sequences and as a result, it is commonly employed for genotyping2. Currently, there are 12 genotypes of MuV circulating around the world (A-N, excluding E and M); with a great deal of diversity3. Mumps is a vaccine preventable disease, and its efficacy is determined by the decrease in the number of cases after administration of two doses of Measles-Mumps-Rubella (MMR) vaccine4. Despite widespread vaccination, resurgence of mumps cases and outbreaks continued to occur5. This might be due to decreased immunity or due to loss of neutralizing epitopes between the circulating and vaccine strains. The decision by the Government of India to exclude mumps from the Universal Immunization programme was influenced by several factors like perception of mumps as a seemingly insignificant public health concern, lack of published data on the community burden of mumps and economic considerations related to vaccine costs. Though it was considered a non-significant problem but several regions in India viz., Karnataka, Maharashtra, Tamil Nadu, and Punjab have reported mumps outbreaks with circulation of C and G genotypes6-9. Such outbreaks and resurgence of mumps have raised queries regarding the effectiveness of currently administered MMR vaccines in private healthcare settings because strains included in the vaccines are of Genotype A (Jeryl Lynn) and Genotype N (L Zagreb). These vaccines are used as an optional vaccine6,10 adding concerns about their ability to prevent outbreaks. Whole genome sequencing (WGS) may be helpful and can aid in identifying genetic variations of the circulating strains, providing a more comprehensive understanding of disease epidemiology.
As per author knowledge to date, 13 MuV complete genome sequences from India are available in the Gen Bank database. However, representative whole genome data from Assam, northeast India, are not available in public repositories. Therefore, the present investigation attempted to study the complete genome sequences of a representative MuV genotype C from Dibrugarh, Assam, and compare them with other reference genomes to identify the genetic variations of the circulating strains.
Material & Methods
The present study was undertaken at Regional Viral Research and Diagnostics Laboratory, ICMR-Regional Medical Research Centre-NE region, Dibrugarh, Assam after obtaining approval from the Institutional Human Ethics Committee.
Study design
A representative sample of MuV Genotype C detected by sequencing the SH gene from our published study (GenBank Accession No. OK335130)11 was selected for WGS analysis considering the low CT value and high viral load. RNA was extracted and quantified for the Whole genome sequencing using the Nanodrop (Thermo Fisher Scientific, Massachusetts, USA). The library preparation was done using the NEBNext Ultra II Directional Library Prep Kit (New England Biolabs, Ipswich, MA, USA) and sequencing was carried out using the Illumina MiSeq platform (https://www.illumina.com/systems/miseq.html). An average of 5M paired-end reads were generated. Paired-end raw reads with a fastq format were subjected to quality control check using the NGSQC toolkit12. Stringent criteria of 70:30 was used to obtain high-quality filtered reads wherein more than 70 per cent high quality (HQ) bases, each having phred scores >30. About 99 per cent of the reads were left after filtering and considered for building viral assembly. HQ filtered paired-end libraries of the samples were used to construct assembly using RNAviralSPAdes v3.15.3 (https://github.com/ablab/spades), which resulted in a single scaffold covering the whole genome of MuV. Genome annotation was done based on WHO (World Health Organization) reference genome of mumps virus, genotype C (Accession no EU370206, MuVi/Zagreb.HRV/39.98). It was submitted to GenBank (accession number OR888547).
Genome wide mutational profiling
The complete genome sequence of Assam MuV Genotype C was aligned and analyzed with the WHO reference genome of Genotype C, vaccine strain of Genotype A (AF338106) and N (AY685920)7,11 and all complete MuV genomes belonging to genotype C from India. Pair-wise nucleotide and amino acid distance of Assam MuV was also calculated. To determine N-glycosylation sites, the server NetNGlyc 1.0 was used13.
Phylogenetic analysis
For phylogenetic analysis, the whole genome sequences of MuV from different parts of India, along with 27 WHO reference strains of MuV, eight vaccine strains and the studied Assam MuV, were selected. Sequences from worldwide representing the 12 MuV genotypes were also included. The tree was constructed using MEGA XI (Molecular Evolutionary Genetics Analysis version 11)14 software using the maximum Likelihood method with General Time Reversible and Gamma distribution models and 1000 bootstrap replicates.
Epitope screening
Considering the selection criteria as: organism: Mumps orthorubulavirus (IEDB ID: 2560602), antigen: HN (IEDB ID: P11235) and F (IEDB ID: P11236), host: Homo sapiens, epitope: any, the Immune Epitope Database (IEDB) (http://tools.iedb.org/conservancy/) was used to identify experimentally determined MuV epitopes available in the database. During the search, no restriction to assay and MHC was applied. Identified epitopes of HN and F protein were mapped against the Assam MuV to analyze their conservancy with the Jeryl Lynn and L Zagreb vaccine strain.
Results
The whole genome sequencing of the Assam MuV genotype C generated 15376 bp in length, which covers the entire MuV genome with protein-coding genes in order. The mutational analysis of the Assam MuV with WHO reference strain for genotype C, vaccine strain Jeryl lynn (Genotype A) and L Zagreb (Genotype N) are illustrated in Table.
| Genes | With WHO reference strain of genotype C (EU370206) (PND/PAD) % |
With genotype A vaccine strain (Jeryl Lynn) (AF338106) (PND/PAD) % |
With genotype N vaccine strain (L-Zagreb) (AY685920) (PND/PAD) % |
|---|---|---|---|
| Nucleoprotein |
I58V, G217R, Q472P, H528P (2.14/0.7) |
I58V, G305D, V455A, Q456H, L457A, T465N, M492I, N495H, L506I, H528P, V529G, L542S (5.2/2.2) | I58V, G305D, H453N, Q456H, M506I, H528P (3.8/1.1) |
| Phosphoprotein |
A45V, I57T, G73S, A159S, A331V (2.4/1.3) |
T27A, S39T, L41I, A45V, I57T, H59Y, H64N, T69S, G73S, V76A, S78P, I80M, P83S, A86G, N88T, T108S, A159S, L192P, S311G, K313E, S326P, A331V, T334P, S336P, Q345R (6.5/6.8) | A45V, I57T, H64N, G73S, K77R, I80M, V106I, T108S, T159S, P161A, L177S, A331V (4/3.2) |
| Matrix | S66A, H104Q, S315T (3.6/0.8) | V82I, H104Q, R138K, Y140H, I207V, R373S (6.7/1.6) | I26V, P79Q, V82I, H104Q, R138K, E228D (4.5/1.6) |
| Fusion | P5L, I125V, Q149R, V278I, K322R, F391S (2.1/1.1) | N2K, P5L, I7T, Y11F, I13V, T24I, V49I, L95P, S177N, R318S, K322R, M331I, T345S, S409A, T431A, F477V, G480N, S489T, I498M (6.1/1.5) |
S5L, I151V, T184S, F195S, V273I, K322R, L475Q, I498M (4/3.7) |
| Small hydrophobic | P7L, V26I, V29I, A42V, H44Y, P48L, S49F (5.7/1.3) |
P7L, V14L, V26I, T28V, L30S, N33T, Y40H, A42V, F48L, S49F, G52S (14.9/2.4) |
P7L, V26I, I28V, L30S, A42V, H44Y, F48L (6.9/1.4) |
|
Hemagglutinin- Neuraminidase |
N12D, A13T, V20G, V23A, D25N, D53G, I118F, N121S, T275I, I287V, S351P, W362L, V375I, H385N, I452V, I473T (3.9/2.7) | I8T, M9I, N12D, A13T, V20G, V21I, G25N, I44T, I56V, R76K, A80T, V81M, I118F, N121S, R122K, N123K, V125A, T130I, I135V, A228V, T275I, I279T, I287V, L336S, S351P, E356D, S372N, V375I, H464N, I473T, V474A, R490S, A577T (8.1/6.4) |
N12D, A13T, V20G, V23A, D25N, N51T, R76K, N80T, I118F, V125A, T130I, I259V, T275I, S351P, V375I, I452V (4.3/2.8) |
| Large | S133L, S347T, N650H, A721S, R916K, I983V, F1715Y, N1729S, K2035R, R2046C, T2051I, I2180M, E2243D, T2247I (2.6/0.6) | A100S, T155A, Y158C, S227N, V272I, I291V, I310V, R485K, N650H, D1210N, I1329V, L1398F, M1468V, Y1661H, D1749E, N1874S, I1876V, V1968I, K2035R, I2043V, S2085P, A2113S, G2241D, E2243D, I2245V, V2247I (6/1.1) | A100S, Y158C, S650H, V695I, T796S, E868D, F949L, F1214S, V1673I, T1750A, Y1806S, I2043V, R2046C, E2243D, I2245V (3.7/0.7) |
WHO, World Health Organization; PND, percentage nucleotide distance; PAD, percentage amino acid distance
Whole genome analysis
The genetic diversity of Assam MuV with Jeryl Lynn and L Zagreb vaccine strains was found to be 6.8 and 4.2 per cent, respectively. Notably, this aligns with the genetic diversity detected among all the whole genome sequences of MuV across various regions of India which ranges from 6.7 to 6.8 per cent when compared to the Jeryl Lynn vaccine strain and from 4 to 4.2 per cent when compared to the L Zagreb vaccine strain. Variation analysis along with percentage nucleotide and amino acid distance of the seven encoding genes of Assam MuV with that of reference genome and two vaccine strains are shown in Table. Nucleoprotein gene analysis showed four amino acid substitutions, which were consistent with those of other Indian isolates. A449V in the Osmanabad 2012 isolates and T531A substitutions present in the Pune 2012 isolates were absent in Assam MuV. Antigenic site analysis (412-475, 475-549) revealed Assam MuV mutations (Q472P, H528P) in the C-terminal region. Analysis of the phosphoprotein gene revealed four amino acid substitutions when compared with the reference genome and analysis with Jeryl Lynn and L Zagreb vaccine strains revealed 25 and 12 substitutions, respectively which are demonstrated in Table. Matrix gene analysis showed three amino acid substitutions S66A, H104Q, S315T in the Assam MuV, which were also present in the Pune-2008, Chennai-2012 and Kushinagar-2013 isolates. Additional substitution at position I40V, T70A, K107R present in Pune Kushinagar isolates were not present in Assam MuV. Comparison with the two vaccine strains revealed six amino acid mutations among which H104Q was common with the Assam MuV. Analysis of the fusion gene revealed substitutions with reference and the vaccine strains. The position 98-102 with Arg-Arg-His-Lys-Arg, also called as the cleavage site were found to be conserved in Assam MuV. Six glycosylation sites present in the F protein at positions 73–75, 182–184, 352–354, 427–429, 433–435, and 457–459 were also present in the Assam MuV. SH gene encodes 316 amino acids, and analysis of the SH gene of Assam MuV with that of the reference genome identified seven substitutions; an analysis with the vaccine strains is shown in Table. Two substitutions at position F12I and A37S in Pune 1986 isolates and at positions I32V and T36S in Pune 2012 isolates were absent in the Assam MuV.
The hemagglutinin-Neuraminidase gene encoding 582 amino acids analyses revealed 16 substitutions compared to the reference genome. Receptor binding site at position 240-245 (NRKSCS) and leucine-zipper, neuraminidase motif at position 405- 410 (GAEGRV) are well conserved in the Assam MuV like other Indian isolates of genotype C. Antigenic sections (265-288, 329-340, 352-360) were preserved, but T275I and I287V substitutions occurred upon comparison with the reference genome which were also present in Pune and Osmanabad isolates. Subsequently, a comparison of the antigenic sites with the Jeryl Lynn vaccine strain revealed three substitutions (T275I, I279T and I287V) and with L Zagreb vaccine strain revealed a single substitution (T275I). Positions 335, 354, 356, 360, 464, and 466 of HN protein have been documented to be associated with neurovirulence15. However, positions showed no mutations in Assam, indicating a lack of neurovirulence. Assam had eight N-linked glycosylation sites (127–129, 284–286, 329–331, 400–402, 448–450, 464–466, 507–509, and 514–516), one more than other Indian isolates, with a mutation at position 12 where asparagine was substituted with aspartic acid affecting glycosylation. Analysis of the large gene with 6925 nucleotides long encodes a putative protein of 2261 amino acids excluding the poly-A tract. The polymerase catalytic site at position 778-780, RNA template contact region at position 555-643 and ATP binding site at position 1814-1818 as described by Jin and co-worker16 are conserved in Assam MuV, like other Indian isolates except Pune 2008 and 2012, isolates where there is a difference at position 581 and 621.
Phylogenetic analysis
Examining the entire genomes through phylogenetic analysis indicated that the Assam MuV under investigation forms a monophyletic clade closely associated with genotype C, clearly distinguishing itself from the two vaccine strains, Jeryl Lynn (genotype A) and L Zagreb (genotype C). These strains are effectively grouped within their designated genotypes, namely genotype A and N. Thus, the phylogenetic analysis clearly shows that Assam MuV under study is of genotype C (Fig. 1).
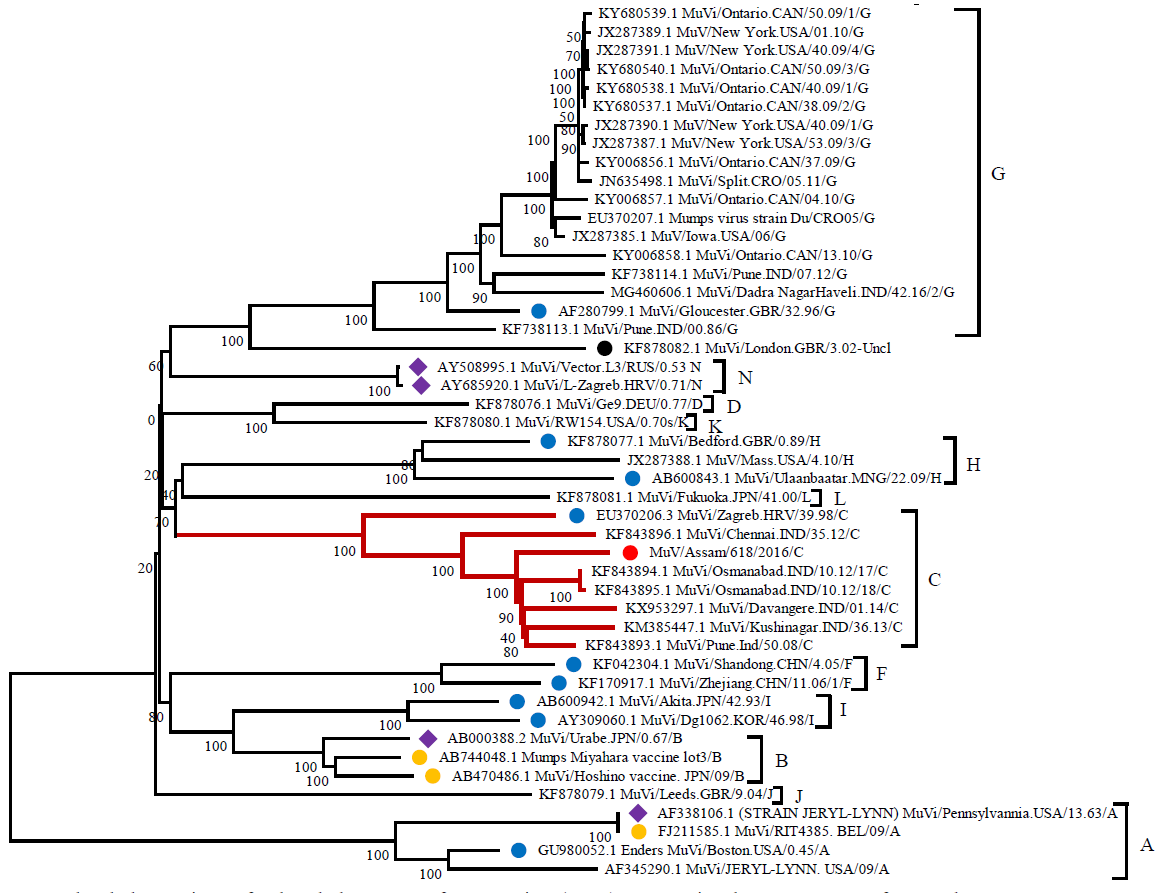
- The phylogenetic tree for the whole genome of mumps virus (MuV) representing the 12 genotypes of MuV. The Assam MuV genotype C is represented with red dot. The WHO (world health organization) reference strains are labeled with blue dot, the vaccine strains are labelled with yellow dot and the strains representing both the WHO reference strain and the vaccine strain are labeled in purple diamond. The sequences are named as follows: Accession number followed by isolate/strain name as in GenBank. The tree was generated using MEGA XI software14.
Screening for conservancy of epitopes
As per author knowledge to date, there are 12 experimentally determined T-cell epitopes for HN protein and 22 epitopes for F protein of MuV in the IEDB. However, no B-cell epitope information is available in the database. The identified T-cell epitopes were aligned against the Assam MuV HN and F protein sequences to check for their occurrence. Subsequent comparisons were made with both vaccine strains, and a percentage conservancy analysis was conducted. The examination of each epitope for both proteins unveiled a high degree of conservancy, with minimal variation observed in certain epitopes, as illustrated in Figure 2.

- Conservancy analysis of Assam MuV T-cell epitopes that were mapped with experimentally determined T-cell epitopes of MuV reported in IEDB (A) HN protein and (B) F protein and further compared against both the vaccine strains. IEDB, immune epitope database.
Discussion
Whole genome sequencing has emerged as a standard and effective method for examining and documenting the circulating MuV strains and distinguishing between outbreak and vaccine strains. It allows for in-depth analysis of genetic material by providing valuable insights into epidemiological surveillance, virtually real-time monitoring, mutational analysis, transmission dynamics, and assessing the vaccine’s effectiveness. Owing to the limited studies on genomic epidemiology of MuV strains, particularly in northeast India, this research focuses on the genetic characterization of circulating MuV genotype C in the Dibrugarh district of Assam.
The two surface glycoproteins, HN, and F protein, generated on the surface of MuV virus, play a crucial role in facilitating virus entry into host cells and disseminating newly formed virions17-20. The HN protein is a primary target for the immune response during mumps infection and vaccination21. Changes (T275I, I279T and I287V) that have occurred in the immune-dominant HN protein when compared to the reference and vaccine strain might explain the incidence of MuV outbreaks in the population or resurgence of Mumps in those that have received vaccinations. As the antigenic regions are responsible for giving rise to B or T-cell epitopes22, a single substitution at these sites may make the epitopes unrecognizable by neutralizing antibodies, contributing to MuV outbreaks. Threonine, which is a small, neutral, uncharged, polar amino acid at position 275 substituted by isoleucine, which is large, hydrophobic, uncharged, and nonpolar may result in loss of hydrogen bond formation between the amino acids as threonine has both hydrogen bond donor and acceptor atoms, but isoleucine is neither a hydrogen bond donor nor acceptor. Again, isoleucine at position 279 substituted with threonine and at position 287 with valine, which is a medium, hydrophobic, uncharged, non-polar amino acid. It was observed that the large amino acid was replaced at both positions by smaller ones, which might result in structural changes. These changes have also been predicted to result in differences of epitopes leading to mismatch of CD4+ and CD8+ responses and hence failure to recall neutralizing antibody23. The function of the F protein is promoted by the first 19 amino acids, which function as a signal peptide and varies with strains24-25. The P gene encodes 391 amino acids, including V protein, after the insertion of two G residues at position 437-438 in the coding region of P, which explains the only mechanism for the generation of P protein26.
Phylogenetic analysis indicated that the examined strain was associated with genotype C and exhibited similarity to MuV strains from different regions of India. When Assam MuV was genetically characterized against two vaccine strains, notable nucleotide and amino acid variations emerged across all seven encoding genes of MuV. These findings prompted the utilization of freely accessible IEDB database to screen the experimentally determined epitopes by IEDB for conservancy analysis. Although, IEDB did not show many discrepancies within the regions of the mapped epitopes, the genetic variations that occurred in different coding genes may lead to the emergence of new candidate epitopes.
The data presented in this study represents a significant step in the field of MuV genetic epidemiology, not only within India but also globally. By genetically characterizing the circulating MuV genotype C in the Dibrugarh district of Assam, this research fills important gaps in our understanding of how mumps viruses evolve and spread by providing valuable insights into the genetic features of MuV and how they mutate and change over time, particularly in regions where such studies have been limited, like Northeast India. Furthermore, by monitoring these genetic changes, researchers can better assess the effectiveness of mumps vaccines. Vaccines are designed to focus on particular virus variations, therefore, genetic shifts in the virus could potentially impact the vaccine’s effectiveness in providing protection. Understanding these changes allows public health officials to adapt vaccination strategies accordingly, ensuring that vaccines remain effective against the disease. The findings of this study have important implications for both scientific understanding and public health practice by devising strategies against the evolving pathogen.
Acknowledgment
None.
Financial support & sponsorship
The study received financial support from Regional VRDL-ICMR, NE Region, a scheme under the Department of Health Research, Ministry of Health and Family Welfare, Government of India (VRDL no. R-15012/31/2020-HR-VRDL).
Conflicts of Interest
None.
Use of artificial intelligence (AI)-assisted technology for manuscript preparation
The authors confirm that there was no use of AI-assisted technology for assisting in the writing of the manuscript and no images were manipulated using AI.
References
- Molecular biology, pathogenesis and pathology of mumps virus. J Pathol. 2015;235:242-52.
- [Google Scholar]
- Circulation of two mumps virus genotypes in an unimmunized population in India. J Med Virol. 2013;85:1426-32.
- [Google Scholar]
- Summary of notifiable diseases---United states, 2005. MMWR Morb Mortal Wkly Rep. 2007;54:1-92.
- [Google Scholar]
- Prevention of measles, rubella, congenital rubella syndrome, and mumps, 2013: summary recommendations of the Advisory Committee on Immunization Practices (ACIP) MMWR Recomm Rep. 2013;62:1-34.
- [Google Scholar]
- Unnoticeable mumps infection in India: Does MMR vaccine protect against circulating mumps virus genotype C? WA SET. 2012;6:1365-71.
- [Google Scholar]
- Genotyping and subtyping of mumps virus isolates from the Indian subcontinent. Arch Virol. 2013;158:2359-63.
- [Google Scholar]
- Characterisation of mumps virus genotype C among patients with mumps in India. Indian J Med Microbiol. 2013;31:290-2.
- [Google Scholar]
- Mumps disease outbreak in Davangere district of Karnataka, India. Indian J Med Microbiol. 2015;33:378-82.
- [Google Scholar]
- Co-circulation of two mumps virus genotypes in Assam, India. Virus Genes. 2023;59:515-23.
- [Google Scholar]
- NGS QC toolkit: a toolkit for quality control of next generation sequencing data. PLoS One. 2012;7:e30619.
- [Google Scholar]
- Prediction of glycosylation across the human proteome and the correlation to protein function. Pac Symp Biocomput. 2002;7:310-22.
- [Google Scholar]
- MEGA11: Molecular evolutionary genetics analysis version 11. Mol Biol Evol. 2021;38:3022-27.
- [Google Scholar]
- Characterization of mumps virus strains with varying neurovirulence. Scand J Infect Dis. 2005;37:330-7.
- [Google Scholar]
- The genomic sequence of a contemporary wild-type mumps virus strain. Virus Res. 2000;70:75-83.
- [Google Scholar]
- Expression of mumps virus glycoproteins in mammalian cells from cloned CDNAs: both F and HN proteins are required for cell fusion. Virology. 1992;187:801-4.
- [Google Scholar]
- Paramyxovirus membrane fusion: lessons from the F and HN atomic structures. Virology. 2006;344:30-7.
- [Google Scholar]
- Trisaccharide containing A2,3-linked sialic acid is a receptor for mumps virus. Proc Natl Acad Sci USA. 2016;113:11579-84.
- [Google Scholar]
- Structural basis for glycan-receptor binding by mumps virus hemagglutinin-neuraminidase. Sci Rep. 2020;10:1589.
- [Google Scholar]
- Localization of a new neutralizing epitope on the mumps virus hemagglutinin-neuraminidase protein. Virus Res. 2001;74:133-7.
- [Google Scholar]
- Are cases of mumps in vaccinated patients attributable to mismatches in both vaccine T-cell and B-cell epitopes? An immunoinformatic analysis. Hum. Vaccines Immunother. 2014;10:290-300.
- [Google Scholar]
- Molecular cloning and characterization of six genes, determination of gene order and intergenic sequences and leader sequence of mumps virus. J Gen Virol. 1988;69:2893-900.
- [Google Scholar]
- Molecular characterisation of two mumps virus genotypes circulating during an epidemic in Lithuania from 1998 to 2000. Arch Virol. 2002;147:243-53.
- [Google Scholar]
- Strain-variable editing during transcription of the P gene of mumps virus may lead to the generation of non-structural proteins NS1 (V) and NS2. J Gen Virol. 1990;71:1555-60.
- [Google Scholar]






