Translate this page into:
Molecular characterization of influenza A(H1N1)pdm09 viruses circulating at various geographical locations in India, 2017
VRDL Team: Vegad M, Byramjee Jeejeebhoy Medical College, Ahmedabad; Dwivedi B, Sabat J, Regional Medical Research Centre, Bhubaneswar; Sarmah K, Regional Medical Research Centre, Dibrugarh; Sharma A, Gauhati Medical College, Guwahati; Sharma P, Tiwari J, Sawai Man Singh Medical College, Jaipur; Jain A, Prakash S, King George's Medical University, Lucknow; Jagadesh A, Krishnan A, Manipal Institute of Virology, Manipal Academy of Higher Education (Deemed to be University), Manipal
For correspondence: Dr Mandeep Chadha, ICMR-National Institute of Virology, 20A Dr Ambedkar Road, Pune 411 001, Maharashtra, India e-mail: mscniv@gmail.com
-
Received: ,
This is an open access journal, and articles are distributed under the terms of the Creative Commons Attribution-NonCommercial-ShareAlike 4.0 License, which allows others to remix, tweak, and build upon the work non-commercially, as long as appropriate credit is given and the new creations are licensed under the identical terms.
This article was originally published by Wolters Kluwer - Medknow and was migrated to Scientific Scholar after the change of Publisher.
Abstract
Background & objectives:
Influenza virological surveillance is an essential tool for the early detection of novel genetic variants of epidemiologic and clinical significance. This study was aimed to genetically characterize A(H1N1)pdm09 virus circulating in 2017 and to compare it with the global data.
Methods:
The regional/State Viral Research and Diagnostic Laboratories (VRDLs) provided influenza diagnosis for referred clinical samples and shared influenza A(H1N1)pdm09 positives with the Indian Council of Medical Research-National Institute of Virology (ICMR-NIV), Pune, India, for hemagglutinin (HA) gene phylogenetic analysis. Sites at Manipal, Jaipur and Dibrugarh performed the sequencing and shared the sequence data for analysis. The antiviral susceptibility of influenza viruses was assessed for known molecular marker H275Y at the ICMR-NIV, Pune.
Results:
All the eight VRDLs had well-established influenza diagnostic facilities and showed increased activity of influenza A(H1N1)pdm09 during 2017. Phylogenetic analysis showed that the viruses from the different regions of the country were similar to A/Michigan/45/2015 strain which was the 2017-2018 recommended vaccine strain and were clustered with the globally circulating clade 6B.1 with signature mutations S84N, S162N and I216T. The clade 6B.1 showed further subgrouping with additional mutations S74R, S164T and I295V; however, there was no significant association between the presence of these mutations and severity of disease due to influenza. All the study viruses were sensitive to oseltamivir.
Interpretation & conclusions:
During the study period, all the study sites reported globally circulating A/Michigan/45/2015 vaccine strain of influenza A(H1N1)pdm09 viruses and remained sensitive to oseltamivir. Further genetic and antigenic characterization of influenza viruses is recommended to address public health concerns.
Keywords
H275Y
haemagglutinin protein
India
influenza A(H1N1)pdm09
Pandemic influenza A(H1N1)2009 virus first appeared in India in May 2009 and thereafter continued to circulate with considerable morbidity and mortality in many parts of the country12345. In India, multisite epidemiological and virological influenza surveillance established previously reported peak influenza activity to be associated with rainy season and a secondary peak in the winter months2. Hemagglutinin (HA) gene sequencing and whole-genome analysis of viruses from three interseasonal upsurges i.e., 2012, 2015 and 2017 have been studied6. The outbreaks of the years 2012, 2015 and 2017 were more widespread across many States such as Rajasthan, Gujarat, Maharashtra, Madhya Pradesh and Telangana7. India has also experienced high mortality associated with influenza A(H1N1)pdm09 virus8.
During the past 10 years, antiviral drugs have provided an important intervention for the treatment and prophylaxis of influenza virus infection. In India, antiviral drugs such as oseltamivir and zanamivir are licensed, and national guidance on their use in clinical management is followed9. The use and stockpiling of antiviral drugs are key components of the pandemic preparedness plans. Surveillance of antiviral susceptibility is, therefore, essential for public health. Initial studies from our group showed that the emergence of neuraminidase inhibitor-resistant viruses was rare1011, and H275Y mutation in neuraminidase (NA) gene was responsible for reducing susceptibility.
This study was undertaken to investigate the HA gene evolution and antiviral drug susceptibility of the 2017 influenza A(H1N1)pdm09 virus and compare it with the Indian, global data, including contemporary influenza vaccine components51213. The study was undertaken at eight Viral Research and Diagnostic Laboratories (VRDLs) across the country established under the Department of Health Research/Indian Council of Medical Research (DHR/ICMR), Government of India, New Delhi, India.
Material & Methods
The samples of suspected influenza patients in category C as defined by the Government of India9 were referred to the VRDLs for diagnosis of influenza. Influenza A(H1N1)pdm09-positive clinical samples were stored at −70°C for future use at the respective VRDLs. Complete HA gene sequencing was done by the VRDLs on randomly selected 10 positive A(H1N1)pdm09 clinical samples representative of severe and mild cases using the WHO sequencing protocol12. Of the eight VRDLs, those of Dibrugarh (365-1650 bp), Jaipur and Manipal carried out sequencing in their respective laboratories and submitted the HA sequences to the ICMR-NIV, Pune. The remaining laboratories submitted the clinical samples tothe ICMR-NIV, Pune, for gene sequencing. Viral RNA was extracted from clinical samples using the Mag Max 96 Kit (Ambion, CA, USA). One-step reverse transcription-polymerase chain reaction (RT-PCR) (Invitrogen Superscript III Platinum Kit, CA, USA) was used to amplify the entire HA gene (~1700 bp) in five overlapping fragments of A(H1N1)pdm09 using the WHO protocol12.
PCR amplicons were subjected to DNA sequencing using Big Dye Terminator Kit Ver.3.1. (Austin, TX, USA). The expected amplicons for each fragment were visualized on two per cent agarose and purified using Charge switch magnetic beads PCR purification kit (Invitrogen, CA, USA). DNA sequencing was carried out using Big Dye terminator V 3.1 cycle sequencing ready reaction kit (ABI, CA, USA), and unincorporated labelled ddNTPs (dideoxynucleotide triphosphates) were purified using DyeEx 2.0 dye-terminated removal kit (Qiagen, Hilden, Germany). The sequencing was done on ABI 3730 DNA analyzer. The obtained sequence information was edited by Seqscape V2.5 software (Applied Biosystems, USA), and pairwise sequence alignment and phylogeny were performed with MEGA6 program13.
The ICMR-NIV, Pune (Apex laboratory) collated all the data and performed quality checks on the raw sequence data received from the three laboratories. A phylogenetic tree was constructed based on the sequences of this study together with the sequences from India and neighbouring countries available from genebank data set and the 2017-2018 WHO recommended vaccine strains14. A neighbour-joining tree was generated using Tamura-Nei best-fit Model for influenza12. Sequence data were deposited in the gene bank, and accession numbers are listed in the Table.
| Strain name | Accession number | Glycosylation site | Amino acid change | Susceptibility to antiviral (oseltamivir) |
|---|---|---|---|---|
| A/India/Dib-1704/2017 | MG271753 | 5 (162,276,287,481,540) | S74R, S164T, I295V | Sensitive |
| A/India/Dib-1572/2017 | MG271752 | 5 (162,276,287,481,540) | S74R, S164T, I295V | Sensitive |
| A/India/Dib-1301/2017 | MG279393 | 5 (162,276,287,481,540) | S74R, S164T, I295V | Sensitive |
| A/India/Dib-1328/2017 | MG271752 | 5 (162,276,287,481,540) | S74R, S164T, I295V | Sensitive |
| A/India/Dib-1737/2017 | MG271754 | 5 (162,276,287,481,540) | S74R, S164T, I295V | Sensitive |
| A/India/Dib-1172/2017 | MF951088 | 5 (162,276,287,481,540) | S74R, I295V | Sensitive |
| A/India/Dib-1745/2017 | MG271754 | 5 (162,276,287,481,540) | S74R, I295V | Sensitive |
| A/India/Guw-20/2017 | MH229485 | 9 | S74R, S164T, I295V | Sensitive |
| A/India/Guw-22/2017 | MH229486 | 9 | S74R, S164T, I295V | Sensitive |
| A/India/Guw-19/2017 | MH229487 | 9 | S74R, I295V | Sensitive |
| A/India/Guw-12/2017 | MH229489 | 9 | S74R, I295V | Sensitive |
| A/India/Guw-30/2017 | MH229488 | 9 | S74R, I295V | Sensitive |
| A/India/Jai-8/2017 | MH333271 | 9 | S74R, I295V | Sensitive |
| A/India/Jai-4/2017 | MH333272 | 9 | S74R, I295V | Sensitive |
| A/India/Jai-13/2017 | MH333273 | 9 | S74R, I295V | Sensitive |
| A/India/Luc-1842492/2017 | MH211344 | 9 | S74R, S164T, I295V | Sensitive |
| A/India/Luc-1842495/2017 | MH211345 | 9 | S74R, S164T, I295V | Sensitive |
| A/India/Luc-1842484/2017 | MH211341 | 9 | S74R, S164T, I295V | Sensitive |
| A/India/Luc-1842489/2017 | MH211342 | 9 | S74R, S164T, I295V | Sensitive |
| A/India/Luc-1842490/2017 | MH211343 | 9 | S74R, S164T, I295V | Sensitive |
| A/India/Ahm-1841326/2017 | MH229460 | 9 | S74R, S164T, I295V | Sensitive |
| A/India/Ahm-1841337/2017 | MH229458 | 9 | S74R, S164T, I295V | Sensitive |
| A/India/Ahm-1841328/2017 | MH229459 | 9 | S74R, S164T, I295V | Sensitive |
| A/India/Bhu-33170/2017 | MH229454 | 9 | S74R, S164T, I295V | Sensitive |
| A/India/Bhu-33201/2017 | MH229456 | 9 | S74R, S164T, I295V | Sensitive |
| A/India/Bhu-33206/2017 | MH229457 | 9 | S74R, S164T, I295V | Sensitive |
| A/India/Bhu-33174/2017 | MH229455 | 9 | S74R, S164T, I295V | Sensitive |
| A/India/Bhu-33144/2017 | MH229453 | 9 | S74R, I295V | Sensitive |
| A/India/Pun-1722287/2017 | MF319590 | 9 | S74R, I295V | Sensitive |
| A/India/Pun-1720775/2017 | MF319572 | 9 | S74R, I295V | Sensitive |
| A/India/Pun-1722256/2017 | MF319588 | 8 (Loss of N-glycosylation) | S74R, I295V | Sensitive |
| A/India/Pun-1722376/2017 | MF319587 | 9 | S74R | Sensitive |
| A/India/Pun-1726441/2017 | MG271886 | 9 | S74R, I295V | Sensitive |
| A/India/Pun-1728697/2017 | MG271900 | 9 | S74R, S164T, I295V | Sensitive |
| A/India/Pun-1727283/2017 | MG271898 | 9 | S74R, S164T, I295V | Sensitive |
| A/India/Pun-1728161/2017 | MG271899 | 9 | S74R, I295V | Sensitive |
| A/India/Man-AF7821/2017 | MG572210 | 9 | S74R, S164T, I295V | Sensitive |
| A/India/Man-AF7638/2017 | MG572213 | 9 | S74R, S164T, I295V | Sensitive |
| A/India/Man-AF9834/2017 | MG572216 | 9 | S74R, S164T, I295V | Sensitive |
| A/India/Man-AF9709/2017 | MG572215 | 9 | S74R, S164T, I295V | Sensitive |
| A/India/Man-AF7736/2017 | MG572209 | 9 | S74R, S164T, I295V | Sensitive |
| A/India/Man-AF7881/2017 | MG572211 | 9 | S74R, S164T, I295V | Sensitive |
| A/India/Man-AF7809/2017 | MG572214 | 9 | S74R, I295V | Sensitive |
| A/India/Man-AF3154/2017 | MG572212 | 8 (Loss of N-glycosylation) | S74R, I295V | Sensitive |
| A/India/Man-AF7729/2017 | MG572208 | 8 (Loss of N-glycosylation) | S74R | Sensitive |
| A/India/Man-AG4823/2017 | MH236898 | 9 | S74R, S164T, I295V | Sensitive |
| A/India/Che-1721549/2017 | MF319575 | 9 | S74R, S164T, I295V | Sensitive |
| A/India/Che-1721030/2017 | MF319568 | 9 | S74R, I295V | Sensitive |
| A/India/Che-1720871/2017 | MF319569 | 9 | S74R, I295V | Sensitive |
| A/India/Che-1721657/2017 | MF319573 | 9 | S74R, I295V | Sensitive |
| A/India/Che-1720872/2017 | MF319571 | 9 | S74R, I295V | Sensitive |
| A/India/Che-1722045/2017 | MF319566 | 9 | S74R, I295V | Sensitive |
| A/India/Che-1722634/2017 | MH794229 | 9 | S74R, I295V | Sensitive |
| A/India/Che-1722637/2017 | MH794228 | 9 | S74R, I295V | Sensitive |
| A/India/Che-1722631/2017 | MH794231 | 9 | S74R, I295V | Sensitive |
| A/India/Che-1722633/2017 | MH794230 | 9 | S74R, I295V | Sensitive |
| A/India/Che-1722639/2017 | MF319562 | 9 | S74R, I295V | Sensitive |
| A/India/Che-1721552/2017 | MF319574 | 9 | S74R, I295V | Sensitive |
| A/India/Che-1722640/2017 | MF319563 | 9 | S74R | Sensitive |
S, serine; R, arginine; T, threonine; I, isoleucine; V, valine
The sequences were also analyzed using the free online tool BII Fluserver (http://flusurver.bii.a-star.edu.sg/). Further, for antiviral susceptibility of the influenza A(H1N1)pdm09, the clinical samples and isolates were tested for the detection of H275Y mutation by allelic discrimination real-time RT-PCR using protocol shared by the National Institute of Health, Thailand15 and isolates by phenotypic fluorescent assay described earlier1011. Minor and major phenotypic outliers were identified based on IC50 cut-off values. The Dibrugarh VDRL tested their study clinical samples for H275Y mutation by allelic discrimination real-time RT-PCR.
Results & Discussion
In 2017, an increased activity of influenza A(H1N1)pdm09 was observed with substantial mortality from most parts of India8. In the present study, analysis of the HA gene-based, 2017 influenza A(H1N1)pdm09 viruses circulating in different regions of India was carried out. Phylogenetic analysis revealed that these were similar to the influenza A/Michigan/45/2015 and were clustered in the globally circulating clade 6B.1 with signature mutations S84N, S162N and I216T (Figure). Though the global frequencies of these mutations were more than 66 per cent, antigenically, the virus remained similar to the A/Michigan/45/2015 strain16. These mutations seem to be fixed after 2016. The clade 6B.1 showed further subgrouping with additional mutations S74R, S164T and I295V, and analysis of clinical data and outcome of the influenza disease showed no significant association between the presence of mutations and severity of disease.
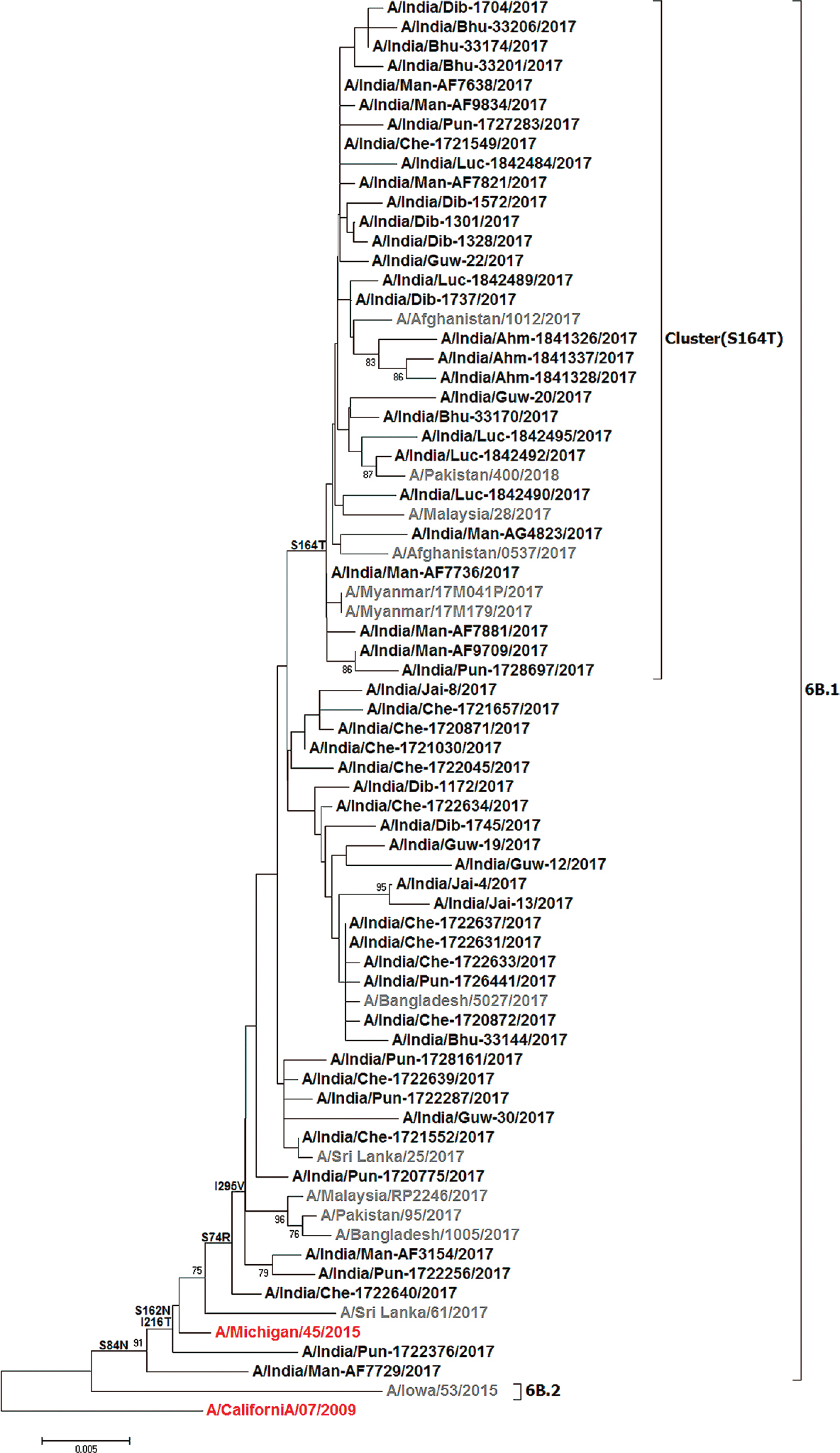
- Phylogenetic analysis of the HA gene of A(H1N1)pdm09 2017 strains from India. The phylogenetic tree was constructed using the MEGA6 software with the neighbour-joining method. The Indian strains are represented in black and global representative strains in grey. The vaccine components are indicated in red; the scale bar indicates the nucleotide substitutions per site.
The accession numbers, amino acid (aa) changes, glycosylation sites and virus susceptibility are summarized in the Table. All viruses were found to be sensitive to oseltamivir with H275. The IC50 values of all the study viruses showed normal inhibition (IC50 range 0.1 to 20 nMols) as compared to that of mutant virus (450-1200 nMols) in phenotypic assay.
Since the emergence of the pandemic influenza A/H1N1 virus in 2009, it has undergone considerable molecular evolution. However, the virus remained antigenically similar to the influenza A/California/07/2009 strain which was the recommended vaccine component from 2009 to 2016. The WHO updated the vaccine components for 2016-2017 northern hemisphere (NH) and 2017 southern hemisphere (SH) from A/California/07/2009 virus to A/Michigan/45/2015 strain, and these viruses were detected globally since 2015-2016 representing Clade 6B.114. During the study period, it was found that the influenza A(H1N1)pdm09 viruses circulating in several parts of India were similar to the globally circulating A/Michigan/45/2016-like viruses which were 2017 SH and 2017-2018 NH vaccine components. Further, these viruses on HA gene phylogeny showed evolution and subgrouping with S74R, S164T and I295V mutations. These mutations were found to be signature mutations of the 2017 strains. It is presumed that these are going to predominate in the future also17. The S164T was the most recent mutation seen in the 2017 strains, and analysis with the global strains showed that about 32 per cent of sequences had this mutation; however, analysis of clinical data and outcome of the influenza disease did not indicate any significant association between the presence of mutation and severity.
The laboratories from Pune, Chennai, Manipal and Dibrugarh had the capacity for virus isolation and isolated 2017 viruses. Thus, these influenza A(H1N1)pdm09 viruses antigenically remained the same as that of A/Michigan/45/2016 strain. The amino acid changes in HA genes S74R and S164T observed in the Indian strains and globally are believed to play a role at the viral oligomerization interfaces, antibody recognition sites and binding small ligands, while I295V also has a probable role in binding ligands (http://flusurver.bii.a-star.edu.sg/).
The previously reported influenza A/California/07/2009-like Indian viruses1213 possess eight glycosylation sites at positions 10, 11, 23, 87, 276, 287, 481 and 540 in HA gene. Notably, three virus isolates in this study from Manipal (2) and Pune (1) had eight glycosylation sites similar to the A/California/07/2009 strain. However, all the other Indian A/Michigan/45/2015-like viruses possessed an amino acid change at S162N that resulted in a gain of N-glycosylation sites at 162 positions in HA gene. Seven partial sequences from Dibrugarh showed five glycosylation sites at amino acid position 162, 276, 287, 481 and 540. Influenza A(H1N1)pdm09 antibodies were directed to each of the two-strain specific (Sa and Sb) and common antigenic sites (Ca and Cb) of the virus HA18. In the present study, A/Michigan-like A(H1N1)pdm09 study viruses showed altered amino acids in the antigenic sites. The sites S157L (singleseq from Jai-8), L161I (singleseq from Luc-1842490) and S164T (28seq) falls in Sa; R205K (singleseq from Pun-1720775) falls in Ca1; while S74R(50) fall in Cb. These antigenic sites could be important for host immune response against influenza A(H1N1)pdm09.
In conclusion, increased activity of influenza A(H1N1)pdm09 was observed in 2017, and A/Michigan/45 2016-like viruses were in circulation in different parts of India. The study also highlighted the capacity of the VRDL network to determine the antiviral susceptibility of circulating viruses which would be helpful in patient and contact management in addition to controlling the outbreaks of influenza. Continued surveillance throughout the country is required for the early detection of genetic changes in the virus and emergence of antiviral-resistant viruses, especially if this occurs in clusters.
Financial support & sponsorship: The study was funded and supported by the Department of Health Research-Indian Council of Medical Research Viral Research and Diagnostic Laboratories (VRDL) and ICMR-National Institute of Virology, Pune.
Conflicts of Interest: None.
References
- Pandemic influenza (H1N1) 2009 is associated with severe disease in India. PLoS One. 2010;5:e10540.
- [Google Scholar]
- Dynamics of influenza seasonality at sub-regional levels in India and implications for vaccination timing. PLoS One. 2015;10:E0124122.
- [Google Scholar]
- Differences in influenza seasonality by latitude, Northern India. Emerg Infect Dis. 2014;20:1723-6.
- [Google Scholar]
- The high frequency of non-aspartic acid residues at HA222 in influenza A(H1N1) 2009 pandemic viruses is associated with mortality during the upsurge of 2015: A molecular and epidemiological study from central India. Epidemiol Infect. 2017;145:2656-65.
- [Google Scholar]
- Molecular and epidemiological analysis of pandemic and post-pandemic influenza A(H1N1)pdm09 virus from central India. J Med Virol. 2018;90:447-55.
- [Google Scholar]
- Influenza A(H1N1)pdm09 outbreak detected in inter-seasonal months during the surveillance of influenza-like illness in Pune, India, 2012-2015. Epidemiol Infect. 2017;145:1898-909.
- [Google Scholar]
- Integrated Disease Surveillance Programme. Seasonal Influenza (H1N1)- State/UT – wise, year-wise number of cases and deaths from 2012- 2019 (till 24th March 2019 2019). Available from: http://idsp.nic.in/showfile.php?lid=3933
- [Google Scholar]
- Seasonal influenza HINI. New Delhi: Directorate General of Health Services. Ministry of Health & Family Welfare, Government of India; Available from: https://ncdc.gov.in/index4.php?lang=1&level=0&linkid=119&lid=276
- Guidelines on categorization of seasonal influenza cases during screening for home isolation, testing, treatment and hospitalization (Revised on 18.10.2016). Available from: https://mohfw.gov.in/sites/default/files/394697031477913837_3.pdf
- Oseltamivir-resistant influenza A(H1N1) pdm09 virus: First reported case from India. WHO South East Asia J Public Health. 2013;2:181-3.
- [Google Scholar]
- Antiviral drug profile of human influenza A & B viruses circulating in India: 2004-2011. Indian J Med Res. 2014;140:244-51.
- [Google Scholar]
- Genetic characterization of the influenza A pandemic (H1N1) 2009 virus isolates from India. PLoS One. 2010;5:e9693.
- [Google Scholar]
- Evolutionary dynamics of the influenza A pandemic (H1N1) 2009 virus with emphasis on Indian isolates: Evidence for adaptive evolution in the HA gene. Infect Genet Evol. 2011;11:997-1005.
- [Google Scholar]
- Recommended composition of influenza virus vaccines for use in the 2017-2018 northern hemisphere influenza. Available from: https://www.who.int/influenza/vaccines/virus/recommendations/2017_18_north/en/
- Regional Office for South-East Asia. Monitoring drug resistance in influenza viruses Report of the Regional Workshop Nonthaburi, Thailand 23-28 August 2010. Available from: http://www.searo.who.int/entity/antimicrobial_resistance/BCT_Reports_HLM-409.pdf
- Seasonal influenza circulation patterns and projections for Sep 2017 to Sep 2018. Available from: https://www.biorxiv.org/content/early/2017/09/26/191676.full.pdf
- Seasonal influenza circulation patterns and projections for Feb 2018 to Feb 2019. Available from: https://www.biorxiv.org/content/early/2018/02/25/271114.full.pdf
- The antigenic structure of the influenza virus A/PR/8/34 hemagglutinin (H1 subtype) Cell. 1982;31:417-27.
- [Google Scholar]






