Translate this page into:
Molecular characterization of hepatitis A virus circulating in Uttar Pradesh, India: A hospital-based study
For correspondence: Dr Amita Jain, Department of Microbiology, King George's Medical University, Lucknow 226 003, Uttar Pradesh, India e-mail: amita602002@yahoo.com
-
Received: ,
This is an open access journal, and articles are distributed under the terms of the Creative Commons Attribution-NonCommercial-ShareAlike 4.0 License, which allows others to remix, tweak, and build upon the work non-commercially, as long as appropriate credit is given and the new creations are licensed under the identical terms.
This article was originally published by Wolters Kluwer - Medknow and was migrated to Scientific Scholar after the change of Publisher.
Abstract
Background & objectives:
Hepatitis A is prevalent worldwide and is among the leading cause of acute viral hepatitis in India. Major geographical differences in endemicity of hepatitis A are closely related to hygienic and sanitary conditions and other indicators of the level of socio-economic development. The present study was aimed to know the seropositivity prevalence and predominant circulating strain of HAV in a north India.
Methods:
Patients with acute viral hepatitis were enrolled. Blood samples were collected over a period of one year from June 2016 to May 2017. Serum samples were tested for anti-immunoglobulin M (IgM) HAV antibodies. The seropositive samples were analyzed for HAV-RNA by real-time reverse transcription-polymerase chain reaction (RT-PCR). Samples detected on molecular assay were subjected to conventional semi-nested RT-PCR for VP1 gene. Further sequencing of amplified RT-PCR products was done, and data were analyzed.
Results:
A total of 1615 patients were enrolled, and serum samples were collected and tested. The male:female ratio was 1.3:1 with a mean age of 24.31±17.02 yr (range 0-83 yr). Among these, 128 (7.93%) were positive for anti-HAV IgM antibodies; 41.63 per cent of seropositive patients were in their childhood or early adolescent age group. Of all seropositive samples, 59 (46.09%) were positive for HAV RNA. Genotyping sequencing of 10 representative strains was carried out, and the circulating genotype was found to be IIIA. The nucleotide sequences showed homology among the strains.
Interpretation & conclusions:
Our results showed that hepatitis A was a common disease in children with IIIA as a circulating genotype in this region. In approximately 50 per cent of cases, HAV RNA could be detected. Higher number of HAV IgM-seropositive cases was observed during monsoon period.
Keywords
AVH
genotype
HAV
hepatitis A
endemic
India
Hepatitis A virus (HAV) is transmitted through faeco-oral route by contaminated water and food. Approximately 1.5 million clinical cases of hepatitis A occur worldwide annually1. The incidence rate is strongly related to socio-economic indicators and access to safe drinking water. Approximately 85 per cent of individuals who are infected with HAV recover fully clinically and biochemically within three months, and nearly, all have complete recovery by six months2. It has been seen that severe manifestations are more common in young adults requiring hospitalization with overall case fatality rate of 0.3 per cent34. HAV is a non-enveloped 27 nm, heat, acid- and ether-resistant RNA virus in the hepatovirus genus of the Picornaviridae family5. Its virion contains four capsid polypeptides, designated as VP1 to VP4, which are cleaved post-translationally from the polyprotein product of approximately 7500 nucleotide genome6. It has single serotype with six genotypes. Three HAV genotypes, I, II and III, divided into subtypes A and B, infect humans. Genotype I is prevalent in Europe and North and South America, and genotype III is endemic in Asia7.
HAV is considered to be endemic in India. According to National Centre for Disease Control, India, HAV is responsible for about 10-30 per cent of acute hepatitis cases in individuals with acute liver failure in India8. The Indian population has shown an upward shift in the average age at the first HAV infection, among the socio-economically developed population resulting in pockets of susceptible populations8. Outbreaks of HAV have been reported, mainly affecting young adults from different parts of the country, e.g., in Delhi, Kerala and Shimla91011. Genotypes I and III of HAV are the predominant strains circulating in India8. The present hospital-based study was aimed to know the seropositivity and predominant circulating strain of HAV in Uttar Pradesh (UP), north India.
Material & Methods
Consecutively, all cases with clinical presentation of acute viral hepatitis (AVH) (WHO case definition)12, referred to the Virology Laboratory, department of Microbiology, King George's Medical University, Lucknow, UP, India, during June 2016-May 2017, were enrolled in this observational study. The protocol was approved by the Institutional Ethics Committee and written informed consent was obtained from each participant. Patients of all age groups and both sexes were included. From each patient, blood sample (5 ml) was collected. Blood samples were centrifuged; serum was collected and tested for anti-HAV immunoglobulin M (IgM) using ELISA kit (DIA Pro Diagnostic Bioprobes Srl., Italy). The remaining sample was stored at −70°C. All anti-HAV IgM positive samples were subjected to HAV real-time reverse transcription-polymerase chain reaction (RT-PCR) assay using a protocol described by Costafreda et al13. For molecular profiling, HAV RNA-positive serum samples were subjected to conventional semi-nested RT-PCR targeting 518 base pair (bp) fragment encompassing the VP1 region which was further subjected to sequencing using a protocol described by Tallo et al14. As our study was focussed on phylogenetic analysis of HAV only, the other faeco-orally transmitted viruses were not tested.
All the amplified products were purified and sequenced using BigDye Terminator Cycle-Sequencing Kit (Applied Biosystems, USA) on ABI 3130 genetic Analyzer (Applied Biosystems, USA). Nucleotide sequences were edited and subjected to GenBank using Basic Local Alignment Search Tool (BLAST) programme (www.ncbi.nlm.nih.gov/BLAST) for comparing with all the available similar sequences. Further, phylogenetic analysis was carried out, and tree was constructed by maximum likelihood method using MEGA7 software with the neighbour-joining method from a Kimura 2-parameter distance matrix15. Genotype was determined using reference sequences belonging to different HAV genotypes.
Results & Discussion
A total of 1615 AVH cases were enrolled over a one-year period. The mean age of the participants was 24.31±17.02 yr (range 0-83 yr) and male:female ratio was 1.3:1. Anti-HAV IgM antibodies seropositivity was 7.9 (128/1615) per cent (Table). The mean age of seropositive cases was 9.3±9.4 yr. About 41 per cent of seropositive patients were in their childhood or early adolescent age group. Of the 128 seropositive samples, HAV RNA was detected in 59 (46.09%) samples. The IgM HAV antibodies percentage positivity in AVH cases referred from Sarawasti (22.2%), Unnao (17%) and Gorakhpur (12.5%) districts was high. The seasonal distribution of enrolled cases showed that AVH occurred throughout the year, though both the number of AVH cases and the HAV IgM seropositive cases increased during monsoon period between June and August (Fig. 1). Of the 59 HAV RNA-positive samples, only 10 high viraemia samples could be amplified for VP1 gene by conventional PCR. (Fig. 2)shows phylogenetic tree of 10 representative HAV strains. The HAV strains with genotype IIIA were circulating in this region showing 92-100 per cent of nucleotide identity in a stretch of 408 bp. Estimates of genetic diversity were conducted on MEGA715 and the analysis among 10 strains showed a total of 36 variable sites among 408 bp. The NCBI BLAST analysis showed 2-4 per cent (8-15 bp) variation from its closest match of Indian (accession no. FJ360733.1) and Norwegian strains (accession no. AJ299464.3). There was no nonsense mutation observed in the sequenced region. The three sequences ID HAV-2, 3 and 5 from Saraswati and Lucknow districts were minimally deviated without any significant difference. The NCBI accession numbers of the submitted HAV sequences were MH929445-MH929454.
| Age (yr) | Total patients tested (n=1615) | Total anti-HAV IgM positive (n=128) | ||||
|---|---|---|---|---|---|---|
| Male | Female | Total tested (%) | Male | Female | Total anti-HAV IgM positive (%) | |
| 0-5 | 143 | 76 | 219 (13.5) | 30 | 30 | 60 (27.39) |
| >5-15 | 220 | 124 | 344 (21.3) | 25 | 24 | 49 (14.24) |
| >15-40 | 383 | 408 | 791 (48.9) | 11 | 6 | 17 (2.14) |
| >40-60 | 136 | 73 | 209 (12.9) | 2 | 0 | 2 (0.95) |
| >60 | 42 | 10 | 52 (3.2) | 0 | 0 | 0 |
| Total | 924 (57.02) | 691 (42.7) | 1615 | 68 (53.12) | 60 (46.87) | 128 (7.9) |
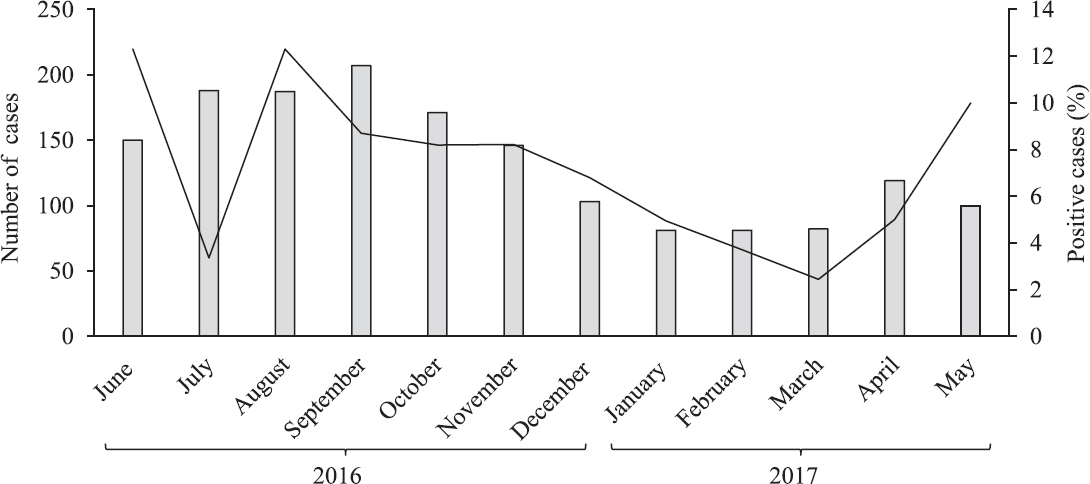
- Seasonal distribution of the total sample tested and confirmed hepatitis A virus case.
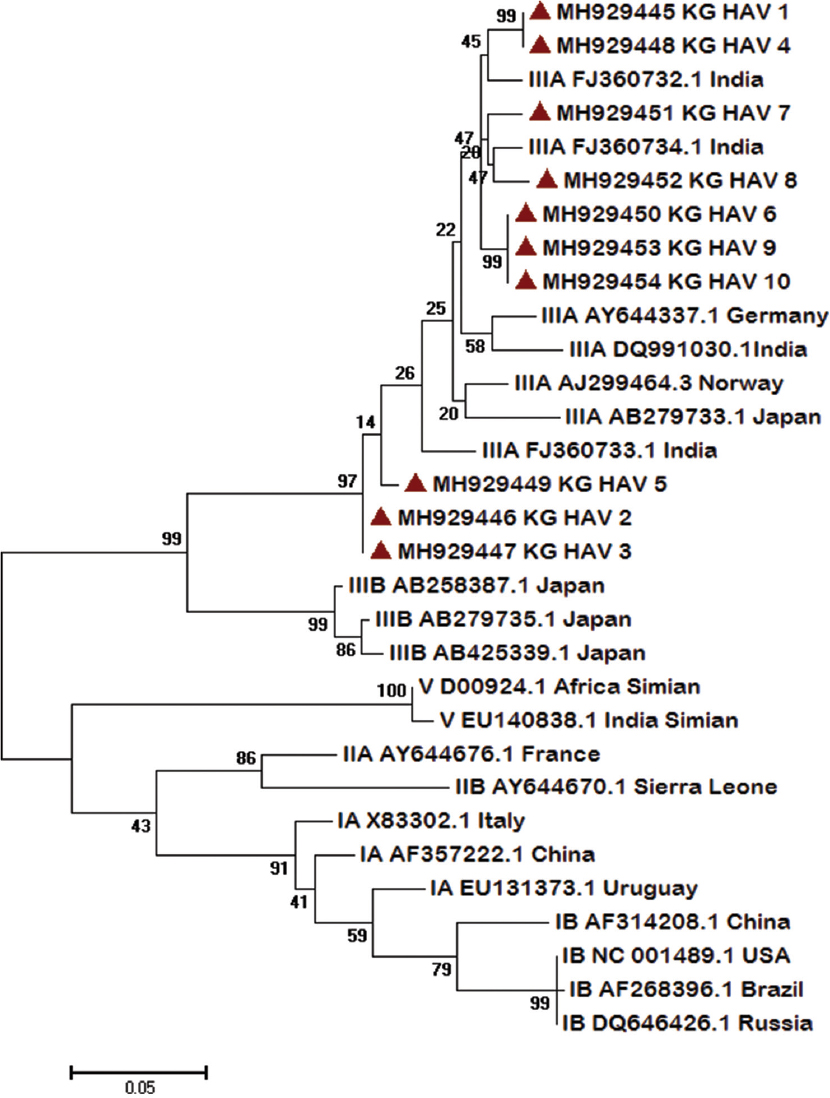
- Molecular phylogenetic analysis of 10 representative hepatitis A virus (HAV) strains by maximum likelihood method. Each strain is labelled by GeneBank accession number followed by country. The strains from this study are marked with (▴). The respective genotypes are mentioned with accession number and country. The evolutionary distances were computed using the Maximum Composite Likelihood method and the tree was drawn to scale, with branch lengths measured in the number of substitutions per site. Evolutionary analyses were conducted in MEGA7.
An earlier study from our centre in 2013 observed high HAV seroprevalence of 26.9 per cent16. The HAV positivity has declined over the time, and this may be due to the continuous effort by the government in improving sanitation and hygiene; however, the need for continuous surveillance can be emphasized looking at the changing pattern. High HAV seropositivity in the early age group depicted HAV endemicity in UP, India. Other studies from Delhi17, Madhya Pradesh18 and Chandigarh19 reported >90 per cent seropositivity in children <15 yr.
HAV RNA positivity among seropositive cases was found to be 46.09 per cent in our study. A similar finding of HAV RNA positivity (47.7%) was reported from Chandigarh in a hospital-based study on 1334 AVH participants19. The molecular detection of HAV RNA ranged from 39.7 per cent in a hospital-based study to 63.2 per cent in outbreak investigations111719.
Peaks in total AVH cases admitted and IgM HAV seropositivity occurred during the monsoon season (June-August). This pattern of upsurge of HAV cases in monsoon has been reported in earlier studies4 and suggests a possibility related to contamination of drinking water during periods of heavy rain. Since the study was done on referred samples, it could not represent true picture as far as district-wise positivity was concerned. More studies on larger sample sizes may be conducted. Moreover, HAV was not detected in several districts; this may be because of less number/adult samples.
Molecular epidemiology of HAV is important to understand the strains circulating in various geographical regions and tracing the source of contamination in an outbreak situation20. On molecular profiling genotype IIIA was found to be the prevalent circulating genotype in UP. Genotype III has been reported as the predominant genotype (70%) followed by genotype IA (30%) from Delhi, north India17. Co-circulation and co-infections with subgenotypes IIIA and IB have been reported from Pune, western India21. HAV genotype IIIA has been reported as an aetiological agent of various other waterborne outbreaks from northern, southern and western India8. The genetic diversity in VP1 gene of this region (2-4%) suggests diversity in circulating genotype IIIA in India and requires further analysis of other gene targets. This study of the mutational analysis of HAV RNA from different parts of the country would determine the characteristics and source of infection and also provide information on trends and transmission pattern.
In conclusion, HAV infection was found common in children in the region. Genotype IIIA was found to be circulating genotype. The mutations at VP1 region warrant further analysis.
Financial support & sponsorship: The financial support received from the Indian Council of Medical Research, New Delhi (Grant 83rd ECM IIA/P9) is acknowledged
Conflicts of Interest: None.
References
- Hepatitis A (WHO/CDS/CSR/EDC/20007). World Health Organization; 2000.
- Hepatitis A virus. In: Knipe D, Howley P, eds. Fields virology (5th ed). Philadelphia, USA: Lippincott Williams Wilkins; 2007. p. :911-47.
- [Google Scholar]
- Viral hepatitis. In: McGill A, Ryan E, Hill D, Solomon T, eds. Hunter's tropical medicine and emerging infectious diseases (9th ed). New York: Saunders Elsevier; 2013. p. :290-305.
- [Google Scholar]
- Surveillance for acute viral hepatitis – United States, 2006. MMWR Surveill Summ. 2008;57:1-24.
- [Google Scholar]
- Avian encephalomyelitis virus is a picornavirus and is most closely related to hepatitis A virus. J Gen Virol. 1999;80(Pt 3):653-62.
- [Google Scholar]
- Genetic relatedness of hepatitis A virus strains recovered from different geographical regions. J Gen Virol. 1992;73(Pt 6):1365-77.
- [Google Scholar]
- National Action Plan Combating Viral Hepatitis in India. New Delhi: Ministry of Health & Family Welfare, Government of India; 2019.
- Epidemiological transition of hepatitis A in India: Issues for vaccination in developing countries. Indian J Med Res. 2008;128:699-704.
- [Google Scholar]
- Molecular characterization of hepatitis A virus from a large outbreak from Kerala, India. Indian J Med Res. 2006;123:760-9.
- [Google Scholar]
- Investigation of a hepatitis A outbreak from Shimla Himachal Pradesh. Indian J Med Res. 2009;130:179-84.
- [Google Scholar]
- WHO-recommended surveillance standard of acute viral hepatitis. Available from: https://wwwwhoint/immunization/monitoring_surveillance/burden/vpd/surveillance_type/passive/hepatitis_standards/en/
- Development, evaluation, and standardization of a real-time TaqMan reverse transcription-PCR assay for quantification of hepatitis A virus in clinical and shellfish samples. Appl Environ Microbiol. 2006;72:3846-55.
- [Google Scholar]
- Sequential changes in hepatitis A virus genotype distribution in Estonia during 1994 to 2001. J Med Virol. 2003;70:187-93.
- [Google Scholar]
- MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 2016;33:1870-4.
- [Google Scholar]
- Prevalence of hepatitis A virus, hepatitis B virus, hepatitis C virus, hepatitis D virus and hepatitis E virus as causes of acute viral hepatitis in North India: A hospital based study. Indian J Med Microbiol. 2013;31:261-5.
- [Google Scholar]
- Increasing trend of acute hepatitis A in North India: Need for identification of high-risk population for vaccination. J Gastroenterol Hepatol. 2006;21:689-93.
- [Google Scholar]
- Circulation of hepatitis A genotype IIIA virus in paediatric patients in central India. Indian J Med Res. 2014;139:940-4.
- [Google Scholar]
- Molecular characterization of hepatitis A virus strains in a tertiary care health set up in North Western India. Indian J Med Res. 2015;141:213-20.
- [Google Scholar]
- Outbreak of infection with hepatitis A virus (HAV) associated with a foodhandler and confirmed by sequence analysis reveals a new HAV genotype IB variant. J Clin Microbiol. 2004;42:2825-8.
- [Google Scholar]
- Exposure of Indian children to hepatitis A virus & vaccination age. Indian J Med Res. 1999;109:11-5.
- [Google Scholar]






