Translate this page into:
Molecular characterization & epidemiology of carbapenem-resistant Acinetobacter baumannii collected across India
For correspondence: Dr Balaji Veeraraghavan, Department of Clinical Microbiology, Christian Medical College, Vellore 632 004, Tamil Nadu, India e-mail: vbalaji@cmcvellore.ac.in
-
Received: ,
This is an open access journal, and articles are distributed under the terms of the Creative Commons Attribution-NonCommercial-ShareAlike 4.0 License, which allows others to remix, tweak, and build upon the work non-commercially, as long as appropriate credit is given and the new creations are licensed under the identical terms.
This article was originally published by Wolters Kluwer - Medknow and was migrated to Scientific Scholar after the change of Publisher.
Abstract
Background & objectives:
Acinetobacter baumannii is an opportunistic pathogen responsible for causing nosocomial infections. A. baumannii develops resistance to various antimicrobial agents including carbapenems, thereby complicating the treatment. This study was performed to characterize the isolates for the presence of various β-lactamases encoding genes and to type the isolates to compare our clones with the existing international clones across five centres in India.
Methods:
A total 75 non-repetitive clinical isolates of A. baumannii from five different centres were included in this study. All the isolates were confirmed as A. baumannii by blaOXA-51-like PCR. Multiplex PCR was performed to identify the presence of extended spectrum β-lactamases (ESBL) and carbapenemases. Multilocus sequence typing was performed to find the sequence type (ST) of the isolates. e-BURST analysis was done to assign each ST into respective clonal complex.
Results:
blaOXA-51-like was present in all the 75 isolates. The predominant Class D carbapenemase was blaOXA-23-like followed by Class B carbapenemase, blaNDM-like. Class A carbapenemase was not observed. blaPER-like was the predominant extended spectrum β-lactamase. ST-848, ST-451 and ST-195 were the most common STs. Eight-novel STs were identified. e-BURST analysis showed that the 75 A. baumannii isolates were clustered into seven clonal complexes and four singletons, of which, clonal complex 208 was the largest.
Interpretation & conclusions:
Most of the isolates were grouped under clonal complex 208 which belongs to the international clonal lineage 2. High occurrence of ST-848 carrying blaOXA-23-like gene suggested that ST-848 could be an emerging lineage spreading carbapenem resistance in India.
Keywords
blaOXA-23-like
carbapenem-resistant Acinetobacter baumannii
CC208
India
multilocus sequence typing
sequence type
Acinetobacter baumannii has emerged as a predominant cause of nosocomial infections across the globe12. A. baumannii can cause a wide range of infections such as ventilator-associated pneumonia, wound infections, urinary tract infections, bloodstream infections and surgical site infections23. Carbapenems are considered to be one of the drugs of choice for treating Acinetobacter infections. However, increased resistance to carbapenem class of antibiotics has been reported worldwide1. Results from studies have reported carbapenem resistance rate of A. baumannii as 40-75 per cent throughout India4.
Carbapenem resistance in A. baumannii is mainly due to Class D and Class B carbapenemases belonging to Ambler's classification of β-lactamases. Although various mechanisms contribute to carbapenem resistance, the majority is due to class D carbapenemases such as blaOXA-23-like, blaOXA-24-like and blaOXA-58-like56. blaOXA-23-like producing A. baumannii responsible for causing outbreaks have been reported from various regions of the world7. Class D carbapenemase in transposons, have the ability to rapidly spread in successful clonal lineages of A. baumannii8.
To control the spread of A. baumannii in the hospital environment, it is necessary to characterize the molecular epidemiology of A. baumannii isolates involved in nosocomial infections9. Multi locus sequence typing (MLST) is highly discriminative and has been successfully applied to various clinically important bacterial pathogens including A. baumannii. MLST offers the possibility of inter-laboratory comparison, thereby providing a powerful tool for epidemiological studies globally10. Published data on the sequence types (STs) of Indian isolates are limited. The aim of the study was to identify the presence of various types of β-lactamases among the carbapenem-resistant isolates and epidemiological typing of A. baumannii by MLST.
Material & Methods
This study included a total of 75 non-repetitive isolates of carbapenem-resistant A. baumannii collected between 2015 and 2017 from five centres across India. These were All India Institute of Medical Sciences (AIIMS, New Delhi), AIIMS trauma centre (New Delhi), Christian Medical College (CMC, Vellore), Jawaharlal Institute of Postgraduate Medical Education & Research (JIPMER, Puducherry) and Postgraduate Institute of Medical Education & Research (PGIMER, Chandigarh). All the study isolates were identified up to the species level as A. baumannii calcoaceticus complex (ABCC) using standard biochemical tests. Further confirmation of ABCC as A. baumannii was performed using blaOXA-51-like PCR which is intrinsic to this species11.
Antimicrobial susceptibility testing: Susceptibility to carbapenem class of antibiotics was determined in the department of Clinical Microbiology, CMC for all the study isolates by Kirby Bauer disc diffusion method and interpreted according to Clinical Laboratory Standard Institute guidelines121314. The antibiotics tested were imipenem (10 μg) and meropenem (10 μg).
Detection of extended spectrum β-lactamase (ESBL) & carbapenemase-encoding genes by multiplex PCR: All the test isolates of A. baumannii were grown overnight on blood agar and genomic DNA was extracted using boiling lysis method15. Conventional multiplex PCR was done for the detection of genes encoding ESBLs such as blaTEM-like, blaSHV-like, blaPER-like and blaVEB-like and carbapenemase genes such as blaGES-like, blaSPM-like, blaIMP-like, blaVIM-like, blaNDM-like, blaKPC-like, blaOXA-48-like and blaSIM-like as reported earlier16. The presence of Class D carbapenemase genes such as blaOXA-23-like, blaOXA-24-like and blaOXA-58-like was also screened by multiplex PCR. The amplicons were visualized in two per cent agarose gel with staining by ethidium bromide. Known positive controls for appropriate genes were used (Courtesy: IHMA, Inc., USA)16. Targeted sequencing was performed to identify the variant of blaTEM-like gene.
Insertion sequence mapping PCR: Mapping PCR was performed for a subset of isolates (n=25) to map the position of insertion sequence, ISAba1 with respect to blaOXA-23-like gene17.
Multilocus sequence typing (MLST): MLST is based on the sequence analysis of seven housekeeping genes and was performed according to Bartual's or oxford scheme10. The seven housekeeping genes included gltA (coding for citrate synthase), gyrB (coding for DNA gyrase subunit B), gdhB (coding for glucose dehydrogenase B), recA (coding for homologous recombination factor), cpn60 (coding for 60 kDa chaperonin), gpi (coding for glucose-6-phosphate isomerase) and rpoD (coding for RNA polymerase 70 factor) were checked. All the seven housekeeping genes were amplified and sequenced using previously described primers10. Each gene sequence was submitted to PubMLST database to find the allelic number and the STs were assigned to each isolate with the seven allelic profiles (http://pubmlst.org/abaumannii/).
e-BURST analysis: e-BURST analysis was performed using the software, e-BURSTv3 (Developed and hosted at The Department of Infectious Disease Epidemiology, Imperial College London) available on the website to assign the STs into respective clonal complexes (http://eburst.mlst.net/) and were defined as single locus (SLVs) and double loci variants (DLVs).
Results
Of the 75 A. baumannii isolates included in this study, 18 were from AIIMS, 22 from AIIMS trauma centre, 25 isolates from CMC, two isolates from JIPMER and eight isolates were from PGIMER. All isolates were resistant to both imipenem and meropenem.
Molecular characterization of ESBL and carbapenemase genes: blaOXA-51-like gene which is intrinsic to A. baumannii, was present in all the 75 isolates (100%). Among the Class D carbapenemases, blaOXA-23-like gene was the most predominant and present in 73 isolates (97%). None of the isolates harboured blaOXA-24-like and blaOXA-58-like genes. Among the Class B carbapenemases, only blaNDM-like gene was found in 13 isolates (17%) whereas other genes such as blaIMP-like, blaVIM-like, blaSPM-like and blaSIM-like were not identified in any of the isolates. None of the isolates had Class A carbapenemase genes like blaKPC-like and blaGES-like. Among the ESBLs, blaPER-like gene was found in 43 isolates (57%). The other ESBL genes like blaSHV-like and blaVEB-like were not identified in any of the isolates tested. blaTEM-like gene was found in five isolates (7%). Distribution of carbapenemase and ESBL genes was similar across all the five study centres. Targeted sequencing revealed that five isolates positive for blaTEM-like belonged to the variant blaTEM-1 gene
Mapping PCR: the ISAba1 element was found upstream of blaOXA-23-like gene in a subset of isolates (n=25) tested.
Multi locus sequence typing: Analysis of multilocus STs of the 75 A. baumannii isolates identified 34 different STs. The predominant ST was ST-848, which was found in 15 isolates (20%), followed by ST-451 in nine isolates (12%), ST-195, in five isolates (7%), ST-218, ST-491 and ST-862 each in three isolates, respectively (4%), ST-208, ST-447, ST-450 and ST-1305 each in two isolates, respectively (3%). Eight novel STs, ST-1500, ST-1501, ST-1502, ST-1503, ST-1504, ST-1505, ST-1506 and ST-1507 were identified in isolates from AIIMS trauma centre. Other less common STs observed in single isolate were ST-229, ST-231, ST-386, ST-391, ST-482, ST-539, ST-620, ST-1051, ST-1114, ST-1223, ST-1289, ST-1306, ST-1307, ST-1308, ST-1335 and ST-1417. ST profiles of the 75 clinical isolates of A. baumannii are summarized in the Table.
| Number of isolates carrying intrinsic OXA gene blaOXA-51-like | Number of isolates carrying acquired OXA gene | Sequence type | Clonal complex | Regional profile | ||
|---|---|---|---|---|---|---|
| blaOXA-23-like | blaOXA-24-like | blaOXA-58-like | ||||
| 15 | 15 | 0 | 0 | 848 | CC208 | Across India |
| 9 | 9 | 0 | 0 | 451 | CC208 | |
| 5 | 5 | 0 | 0 | 195 | CC208 | |
| 3 | 3 | 0 | 0 | 862 | CC862 | |
| 3 | 3 | 0 | 0 | 218 | CC208 | |
| 2 | 2 | 0 | 0 | 208 | CC208 | |
| 1 | 1 | 0 | 0 | 391 | CC447 | South India |
| 1 | 1 | 0 | 0 | 1114 | CC208 | |
| 2 | 2 | 0 | 0 | 1305 | CC208 | |
| 1 | 1 | 0 | 0 | 1306 | CC862 | |
| 1 | 0 | 0 | 0 | 1307 | Singleton | |
| 1 | 1 | 0 | 0 | 1308 | CC862 | |
| 1 | 1 | 0 | 0 | 1335 | Singleton | |
| 1 | 1 | 0 | 0 | 229 | CC229 | North India |
| 1 | 1 | 0 | 0 | 231 | CC231 | |
| 1 | 1 | 0 | 0 | 386 | CC862 | |
| 2 | 2 | 0 | 0 | 447 | CC447 | |
| 2 | 2 | 0 | 0 | 450 | CC208 | |
| 1 | 1 | 0 | 0 | 482 | Singleton | |
| 3 | 3 | 0 | 0 | 491 | CC231 | |
| 1 | 1 | 0 | 0 | 539 | CC208 | |
| 1 | 0 | 0 | 0 | 620 | CC620 | |
| 1 | 1 | 0 | 0 | 1051 | Singleton | |
| 1 | 1 | 0 | 0 | 1223 | CC231 | |
| 1 | 1 | 0 | 0 | 1289 | CC208 | |
| 1 | 1 | 0 | 0 | 1417 | CC208 | |
| 1 | 1 | 0 | 0 | 1500* | CC862 | |
| 1 | 1 | 0 | 0 | 1501* | CC208 | |
| 1 | 1 | 0 | 0 | 1502* | CC208 | |
| 1 | 1 | 0 | 0 | 1503* | CC231 | |
| 1 | 1 | 0 | 0 | 1504* | CC231 | |
| 1 | 1 | 0 | 0 | 1505* | CC862 | |
| 1 | 1 | 0 | 0 | 1506* | CC862 | |
| 1 | 1 | 0 | 0 | 1507* | CC1507 | |
*Novel sequence types, South India-CMC and JIPMER, North India-AIIMS, AIIMS trauma centre and PGIMER. OXA, oxacillinase; CMC, Christian Medical College; JIPMER, Jawaharlal Institute of Postgraduate Medical Education & Research; AIIMS, All India Institute of Medical Sciences; PGIMER, Postgraduate Institute of Medical Education & Research
e-BURST analysis: e-BURST analysis showed that all 75 A. baumannii isolates were clustered into seven clonal complexes (CCs) and four singletons. CC208 was the largest clonal complex and comprised STs 195, 208, 218, 451 and 539 differing in their gpi (SLV), 450, 848, 1114, 1289 and 1305 differing in gpi and gyrB, 1417 in gpi and recA, 1501 in gpi and cpn60 and 1502 in gpi and rpoD (DLV), respectively. The STs 386, 862, 1306, 1308, 1500, 1506 differing in gpi (SLV) and 1505 differing in gpi and cpn60 (DLV) belonged to CC862, while STs 231, 491 differing in gyrB (SLV) whereas 1223 in gyrB and recA, 1503 and 1504 in gpi and gyrB (DLV) belonged to CC231 and STs 391 and 447 differing in gpi and gyrB (DLV) belonged to CC447. Three STs, ST-229, ST-620 and ST-1507 were shown to be the founder of the CCs 229, 620 and 1507, respectively. Four STs, 1307, 1335, 1051 and 482, were found to be singletons (Figure).
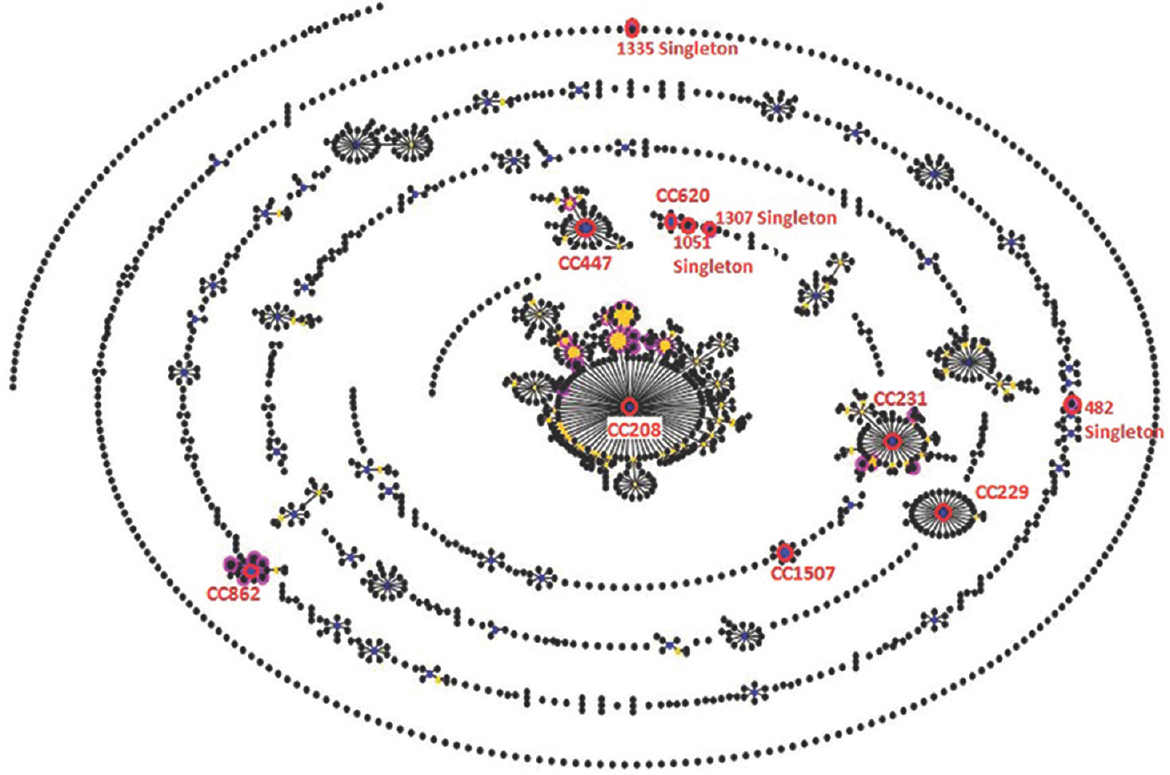
- e-BURST analysis showing clonal complexes and singletons of 34 sequence types of carbapenem-resistant Acinetobacter baumannii. Each circle signifies the sequence type. The size of each circle represents to different number of isolates, with larger sizes corresponding to higher frequency of occurrence.
Clonal complex, CC208, was the predominant and observed across all the five centres in this study. CC862 was observed in AIIMS, AIIMS trauma centre and CMC. CC231 was seen among AIIMS, AIIMS trauma centre and PGIMER isolates. CC447 was observed in AIIMS and CMC centres. CC229, CC620 and CC1507 were observed among AIIMS and AIIMS trauma centres, respectively. Four singletons, 482 from PGIMER, 1051 from AIIMS trauma centre, 1307 and 1335 from CMC were observed.
Discussion
The various molecular typing methods for epidemiological characterization include PCR-based DNA fingerprinting methods such as M13, ERIC and DAF4, restriction enzyme-based methods such as pulsed-field gel electrophoresis and amplified fragment length polymorphism and MLST1819. We followed the Bartual's scheme10, as several studies reported high discriminatory power using Bartual's scheme when compared to Pasteur2021. Carbapenem resistance in A. baumannii is mainly due to Class D oxacillinases. In this study, the predominant OXA group associated with carbapenem resistance across all five centres was blaOXA-23-like16. This was in concordance with other studies where 98 and 81 per cent of the isolates were carbapenem resistant due to blaOXA-23-like gene, respectively2223.
Among the Class B carbapenamases, only blaNDM-like was identified across all the centres except from JIPMER in this study. One study reported blaNDM-like and blaVIM-like as the predominant Class B carbapenemase16 whereas other studies showed blaIMP-like and blaNDM-like as prevalent Class B carbapenemase232425. Saranathan et al26 reported 31 and 15 per cent of blaIMP-like and blaNDM-like, respectively.
Among ESBLs, blaPER-like was the most common and identified across all the four centres whereas other studies showed 54 and 81 per cent of blaPER-like, respectively2226. blaTEM-1 was found only among AIIMS, CMC and PGIMER isolates.
In this study, 34 STs were identified from 75 clinical isolates of A. baumannii. The most common ST identified was ST-848 which was observed among CMC, AIIMS trauma centre, JIPMER and PGIMER isolates. A study from north India reported ST-146, ST-110, ST-69, ST-103, ST-194, ST-108 and ST-188 as the predominant STs and another study from south India showed ST-538, ST-539, ST-103 and ST-576 as the most common STs in their settings2325. None of this study isolates had STs similar to the previously reported STs, except ST-539 which was reported earlier from south India suggesting diverse clonal relatedness23.
The most common clonal complex in this study was CC208, a SLV of the globally disseminated clonal complex CC92 (International clone-II). An earlier study showed CC108 as the predominant clone in India27, while Saranathan et al23 reported CC103 and CC92 as the prevalent clonal complexes in south India. The same investigators also observed the predominance of clonal complexes CC92 followed by CC44726. Although in the current study, none of the isolates belonged to CC92, it was found to be the subgroup founder of CC208.
Molecular epidemiology of clinical isolates of A. baumannii showed eight international clonal lineages (ICL) dominating across the globe. Most common clonal lineages ICL1-ICL3 were initially reported in Europe and the United States, later reported from various countries3. Majority of the carbapenem resistant A. baumannii outbreaks were associated with the ICL2 isolates harbouring blaOXA-23-like carbapenemase gene3. This was in concurrence with this study where majority of the isolates were clustered under CC208 carrying blaOXA-23-like gene.
The main limitation of this study was insufficient number of isolates from two centres (JIPMER & PGIMER) among the five. Characterizing more number of isolates would help to understand the clonal diversity within this region.
In conclusion, our study showed diverse STs among the clinical isolates of A. baumannii across five centres in India. Most of the isolates were grouped under CC208. High occurrence of ST-848 carrying blaOXA-23-like gene suggested that ST-848 might be an emerging lineage spreading carbapenem resistance in India. Further studies with a large number of isolates need to be done to confirm these findings.
Financial support & sponsorship: The authors acknowledge the Indian Council of Medical Research, New Delhi for providing the grant for this research (Ref. No: AMR/TF/55/13ECDII dated 23/10/2013).
Conflicts of Interest: None.
References
- Emergence of carbapenem-resistant Acinetobacter baumannii ST787 in clinical isolates from blood in a tertiary teaching hospital in Northern Taiwan. J Microbiol Immunol Infect. 2017;50:640-5.
- [Google Scholar]
- Molecular epidemiology and characterization of multiple drug-resistant (MDR) clinical isolates of Acinetobacter baumannii. Int J Infect Dis. 2015;41:42-9.
- [Google Scholar]
- Significant spread of extensively drug-resistant Acinetobacter baumannii genotypes of clonal complex 92 among Intensive Care Unit patients in a university hospital in Southern Iran. J Med Microbiol. 2017;66:1656-62.
- [Google Scholar]
- Carbapenem-resistant Acinetobacter baumannii and Enterobacteriaceae in South and Southeast Asia. Clin Microbiol Rev. 2017;30:1-22.
- [Google Scholar]
- Acinetobacter baumannii: Emergence of a successful pathogen. Clin Microbiol Rev. 2008;21:538-82.
- [Google Scholar]
- Emergence of resistance to carbapenems in Acinetobacter baumannii in Europe: Clinical impact and therapeutic options. Int J Antimicrob Agents. 2012;39:105-14.
- [Google Scholar]
- Dissemination of multidrug-resistant Acinetobacter baumannii genotypes carrying bla(OXA-23) collected from hospitals in Rio de Janeiro, Brazil. Int J Antimicrob Agents. 2009;34:25-8.
- [Google Scholar]
- Phenotypic and molecular characterization of clinical isolates of Acinetobacter baumannii isolated from Delhi, India. Ann Clin Microbiol Antimicrob. 2015;14:40.
- [Google Scholar]
- Development of a multilocus sequence typing scheme for characterization of clinical isolates of Acinetobacter baumannii. J Clin Microbiol. 2005;43:4382-90.
- [Google Scholar]
- Identification of Acinetobacter baumannii by detection of the blaOXA-51-like carbapenemase gene intrinsic to this species. J Clin Microbiol. 2006;44:2974-6.
- [Google Scholar]
- Clinical and Laboratory Standards Institute. Performance standards for antimicrobial susceptibility testing; 25th informational supplement. In: CLSI Document M100-S25. Wayne, PA: CLSI; 2015.
- [Google Scholar]
- Clinical and Laboratory Standards Institute. Performance standards for antimicrobial susceptibility testing. In: CLSI Document M100S (26th ed). Wayne, PA: CLSI; 2016.
- [Google Scholar]
- Clinical and Laboratory Standards Institute. Performance standards for antimicrobial susceptibility testing. In: CLSI Document M100-27 (27th ed). Wayne, PA: CLSI; 2017.
- [Google Scholar]
- Heat treatment of bacteria: A simple method of DNA extraction for molecular techniques. Kuwait Med J. 2009;41:117-22.
- [Google Scholar]
- Molecular characterization of invasive carbapenem-resistant Acinetobacter baumannii from a tertiary care hospital in South India. Infect Dis Ther. 2016;5:379-87.
- [Google Scholar]
- The role of ISAba1 in expression of OXA carbapenemase genes in Acinetobacter baumannii. FEMS Microbiol Lett. 2006;258:72-7.
- [Google Scholar]
- Choosing an appropriate bacterial typing technique for epidemiologic studies. Epidemiol Perspect Innov. 2005;2:10.
- [Google Scholar]
- Diversity of multi-drug resistant Acinetobacter baumannii population in a major hospital in Kuwait. Front Microbiol. 2015;6:743.
- [Google Scholar]
- Head-to-head comparison of two multi-locus sequence typing (MLST) schemes for characterization of Acinetobacter baumannii outbreak and sporadic isolates. PLoS One. 2016;11:e0153014.
- [Google Scholar]
- Emergence in Taiwan of novel imipenem-resistant Acinetobacter baumannii ST455 causing bloodstream infection in critical patients. J Microbiol Immunol Infect. 2015;48:588-96.
- [Google Scholar]
- Molecular characterisation of antimicrobial resistance in Pseudomonas aeruginosa and Acinetobacter baumannii during 2014 and 2015 collected across India. Indian J Med Microbiol. 2016;34:433-41.
- [Google Scholar]
- Emergence of carbapenem non-susceptible multidrug resistant Acinetobacter baumannii strains of clonal complexes 103(B) and 92(B) harboring OXA-type carbapenemases and metallo-β-lactamases in Southern India. Microbiol Immunol. 2015;59:277-84.
- [Google Scholar]
- OXA beta-lactamase-mediated carbapenem resistance in Acinetobacter baumannii. Indian J Med Microbiol. 2011;29:269-74.
- [Google Scholar]
- Multi-locus sequence types of Acinetobacter baumanii clinical isolates from India. J Infect Dev Ctries. 2013;7:358-60.
- [Google Scholar]
- Detection of ISAba1 in association with a novel allelic variant of the β-lactamase ADC-82 and class D β-lactamase genes mediating carbapenem resistance among the clinical isolates of MDR A.baumannii. J Med Microbiol. 2017;66:103-11.
- [Google Scholar]
- Spread of carbapenem-resistant Acinetobacter baumannii global clone 2 in Asia and abaR-type resistance Islands. Antimicrob Agents Chemother. 2013;57:5239-46.
- [Google Scholar]






