Translate this page into:
Molecular basis of RhD-negative phenotype in North Indian blood donor population
For correspondence: Dr Dheeraj Khetan, Sanjay Gandhi Postgraduate Institute of Medical Sciences, Raibareli Road, Lucknow 226 014, Uttar Pradesh, India e-mail: dheerajkhetan@gmail.com
-
Received: ,
This article was originally published by Wolters Kluwer - Medknow and was migrated to Scientific Scholar after the change of Publisher.
Abstract
Background & objectives:
RHD gene typing is highly complex due to homology with RHCE genes. Molecular polymorphism of the RHCE and RHD genes have been characterized among various populations, but no studies have been undertaken among Indians. This study was undertaken to assess the genetic basis of RHD-negative phenotype in Indian blood donor population.
Methods:
Sample from a total of 200 phenotypically RhD-negative blood donors were analyzed for presence of RHD gene using polymerase chain reaction (PCR). RHD genotyping was done using three primer sets designed for exons 4 and 10 and one set for identification of pseudo (RHDΨ) gene between introns (int) 3 and 4. Amplified PCR products were analyzed by gel-electrophoresis (XY Loper, Uvitech, Cambridge) and confirmed by nucleotide sequencing (ABI 3730 xl 96 capillary system).
Results:
No PCR product was found in 195/200 (97.5%) of study samples indicating homozygous gene deletion. Of the 5/200 (2.5%) showing RHD gene polymorphisms, 4/200 (2%) were positive for presence of exon 10 only (RHD-CE-D hybrid). RHDΨ gene was not detected in any of the samples tested. One sample showed presence of all three tested regions and was negative for RHDΨ gene.
Interpretation & conclusions:
RHD gene deletion was found to be the most common cause of an RHD-negative phenotype while RHDΨ gene was, reported to be present in up to 39 per cent of various ethnic populations, but was not detected. RHD-CE-D hybrid gene (found in 2.5% individuals) is important for predicting the requirement of Rh prophylaxis during the antenatal period.
Keywords
Blood donor
genetic basis
genotype
phenotype
polymorphism
RH blood group
RHD gene
RhD negative
Molecular biology has been applied extensively in characterizing the genetic basis of blood group systems and for developing clinical diagnostic tools for immune haematology and transfusion medicine1234. There are now 51 antigens within the Rh system and more than 200 alleles for the RHD gene alone.
The strength of an RhD antigen - antibody reaction in individuals may vary due to the absence or weakening of one or few of the 30 different epitopes5 leading to discrepancies in Rh phenotyping using commercial antisera. These individuals are classified as D variants6 which includes partial D and weak D. A negative reaction during serological typing therefore does not necessarily mean absence of RhD antigen. Such individuals may be found to be positive on serological testing with additional antisera or by molecular testing.
Molecular mechanisms producing RhD-negative phenotype differs among various ethnic populations. Deletion of RHD gene is responsible in majority of D-negative Caucasians7, 30 per cent Japanese8 and 10-23 per cent of RhD-negative South African9 population. Another reason may be the presence of a Hybrid allele. Portion of RHCE gene inserted in RHD (Hybrid RHD-CE-D) due to incomplete crossing over may result in a lack of D antigen. Some individuals, particularly of African descent10, have been found to harbour a nonfunctional RHD allele termed as RHD pseudogene (RHDψ) and is caused by a 37 bp insertion leading to the absence of the RhD protein.
A clinical application of the molecular characterization of the RH locus is the assignment of RHD zygosity with certainty. It is important to note that the distinction between apparent RhD-negative and other D variants (including partial D and weak D phenotypes) by serology may be somewhat arbitrary. However, the clinical significance does not encompass: D category of blood transfused to D-negative recipients that may lead to development of antibody. Therefore, in Asian populations, among whom occurrence of D-negative blood is comparatively lesser, identifying such transfusion recipients could reduce the demand for Rh-negative blood11.
It is now widely accepted that molecular analysis is the suggestively the most accurate method of defining the complex RH and other blood group systems. There are now increasing number of clinical settings, where such molecular approaches facilitate preventing blood group incompatibilities, reducing the chances of alloimmunizations and haemolytic transfusion reactions, thus contributing to optimal RBC survival among transfusion-dependent immune disorders.
There is limited data1213 on serological prevalence of D variants (partial D/weak D) using commercially available RhD phenotyping panels and few studies on molecular characterization of suspected partial D samples1415. Kulkarni et al16 have identified hybrid genes in 59/171 (31%) of select (C/E+) subgroup of serological RhD-negative individuals from India. The present study was undertaken to find out prevalence of apparently RhD-negative individuals which may be identified on molecular testing as RhD-positive.
Material & Methods
Study design: This was a prospective study conducted over a period of two years (June 2014 to May 2016), involving a random survey of blood samples that were collected from 200 consecutive RhD sero-negative non-remunerated repeat voluntary blood donors, donating blood at the department of Transfusion Medicine, Sanjay Gandhi Postgraduate Institute of Medical Sciences, Lucknow, after obtaining approval from Institute Ethics Committee and written informed consent from individual participants.
Sample size determination: Since baseline prevalence data on presence of RHD gene in apparently RhD-negative individuals for calculating the sample size was not available from India. Based on published reports from other countries, an average incidence of 20 per cent was assumed and a sample size of 200 was accordingly estimated with the aim of identifying approximately 40 individuals who are serologically RhD-negative samples but have presence of RHD gene.
Blood sample collection: 5 ml of anti-coagulated blood from residual buffy coat (generated during component preparation from whole blood) of RhD-negative blood donors identified during routine serological blood grouping were included in the study.
Serological testing for RhD antigen: Testing for RhD antigen routinely involves initial testing by automated immuno-haematology analyser based on the principle of solid phase red cell adherence assay (Neo, Immucor, USA) using two anti-D antisera [Anti-sera I- Novaclone (IgG and IgM monoclonal, Dominion biological, Nova Scotia, Canada), Anti-sera II- Immucor series 5 (IgG and IgM monoclonal blend, Immucor Norcross, GA, USA)]. Samples identified as RhD-negative on initial testing are further subjected to weak–D testing by column agglutination technology (Biorad Laboratories, GmbH, Switzerland) with the use of IgG monoclonal antisera (ID-DiaClon Anti-D, Bio-Rad, DiaMed GmbH, Switzerland). Donors negative after weak-D testing are considered as RhD-negative.
Serological testing for other Rh antigen: Testing for other Rh blood group antigens (C, c, E and e) was done on all samples by conventional tube technique as per departmental SOP, using commercially available monoclonal (IgM) antisera (C, E, e – Gammaclone, Immucor Norcross GA, USA and c- Series 1, Gammaclone, Immucor Norcross GA, USA).
Molecular testing for RhD antigen: DNA isolation from study samples was done using QiAmp Blood mini kit (QIAGEN, CA) as per manufacturer’s instructions. Primer design and PCR conditions were adopted from earlier published studies9171819. PCR-sequence specific priming was done using three primer sets (Supplementary Table) designed for intron 4, exon 10 and pseudogene in exon 4 (RHDΨ).
| Position | Primer ID | Primer sequence (5′-3′) |
|---|---|---|
| Intron 4 | A1 | ACGATACCCAGTTTGTCT |
| A2 | TGACCCTGAGATGGCTGT | |
| Exon 10 | B1 | TTAAGCAAAAGCATCCAAGA |
| B2 | AATAAATGGTGAGATTCTCCTC | |
| Pseudogene (Exon 4) | C1 | GCCGACACTCACTGCTCTTAC |
| C2 | TCCTGAACCTGCTCTGTGAAGTGC |
Primer set A1, A2 for intron 4: Amplification using this primer set (A1- exon 4; nt 637-654, A2- exon 5; nt 781-798) resulted in a 1200 bp RHCE product and a 600 bp product in RHD-positive individuals. Detection of 1200 bp RHCE product also served as an internal control.
Primer set B1, B2 for exon 10: This primer set was specific for RHD gene sequence (B1- exon 10; nt 1251-nt 1271, B2- exon 10; nt 1421- 1442). Amplification using this primer set specifically amplified a 193 bp product of 3’ un-translated region of exon 10.
Primer set C1, C2 for pseudogene (RHDΨ): This primer set flanked the insertion point of 37 bp sequence within exon 4 (C1- intron 3; nt 36-16, C2- intron 4; nt 174 - nt 197). With this primer pair, the normal RHD allele generated a 381 bp product while RHDΨ generated a 418 bp product.
RHD typing reactions were performed with 250-300 ng of genomic DNA mixed with 10 µl of commercially available PCR master mix (AmpliTaq Gold Fast PCR master Mix, Applied Biosystems, USA), 10 pmol each of forward and reverse primer were added with DEPC treated water (BR Biochem, India) in a final volume of 20 µl. PCR amplification involved a single cycle of 5 min at 95°C and 35 cycles consisting of 45 sec at 94°C, 45 sec at 62°C and 90 sec at 72°C, performed with a Bioer thermal cycler (Bioer Technologies Corp Ltd., China). All PCR reactions were terminated after an 8 min extension at 70°C. DNA from a known RHD-positive sample was run with every run as an external control. Amplified PCR products were analyzed on agarose (2%) gel-electrophoresis (XY Loper, Uvitech, Cambridge), using ethidium bromide stain (BR Biochem, India). Confirmation of amplified product was done by nucleotide sequencing (ABI 3730 XL, 96 capillary system).
Statistical analysis: Data management was done using Microsoft Excel. Frequencies were calculated for different probable genotypes and presented as number and percentages.
Results
Blood samples from a total of 200 blood donors phenotyped as RhD-negative on serological testing were included in the study. Mean age of study population was 31.3±9.04 yr, and majority of them were male (178/200, 89%). ‘B’ was the most common blood group (75/200, 38%) followed by ‘O’ (72/200, 36%), ‘A’ (41/200, 20%) and ‘AB’ (12/200, 06%) in decreasing order of frequency. Majority were Hindus (53%) followed by Muslims (18%), Sikhs (16%), Christians (6%) and others (4%).
Frequency of other Rh antigens (C.c.E.and e): e was found to be the most common antigen (195/200, 97.5%) followed by C (177/200, 88.5%), c (75/200, 37.5%) and E (72/200, 36%) in decreasing order of frequency. Phenotypic frequency and probable RHCE genotype of the study samples (Table I) revealed RHCe/ RHCe (59.4%) as the most common genotype and RHCE/RHCE to be the least common genotypes in our study population.
| Phenotype | Probable genotype | n (%) |
|---|---|---|
| C+, c−, E−, e+ | RHCe/RHCe | 118 (59.4) |
| C+, c+, E+, e+ | RHCE/RHce | 52 (26) |
| RHCe/RHcE | ||
| C−, c+, E+, e+ | RHcE/Rhce | 10 (5.0) |
| C−, c+, E−, e+ | Rhce/RHce | 10 (5.0) |
| C+, c−, E+, e+ | RHCE/RhCe | 5 (2.5) |
| C−, c+, E+, e− | RHcE/RHcE | 3 (1.2) |
| C+, c−, E+, e− | RHCE/RHCE | 2 (0.9) |
Genotyping for RHD: RHD genotyping reaction with the primer pair A1-A2 is shown in Fig. 1, illustrating the pattern of reaction observed for intron 4 region of RHD gene. Complete concordance was observed between 199 RhD sero-negative individuals as no PCR product was identified in these samples. Target sequence in the intron 4 region of RHD gene was identified in only one out of total 200 samples tested.
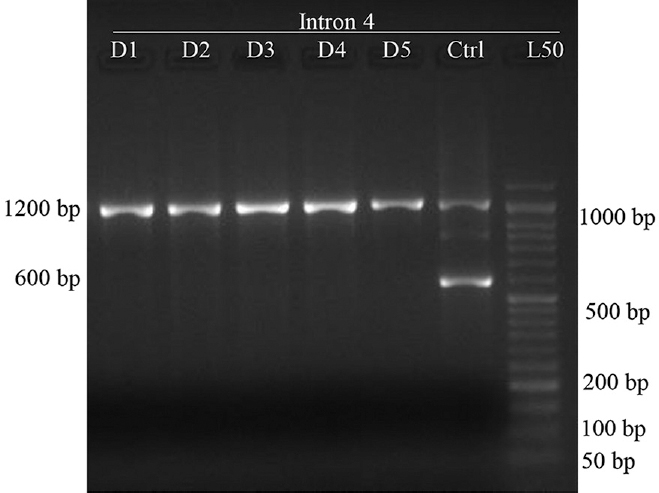
- PCR-sequence specific priming products for intron 4 region of RHD gene (n=200). D1-D5 shows reaction of five donor samples using primer set A1-A2, Ctrl-control sample (DNA from known RhD-positive individual was used as control), L50 - 50 bp DNA ladder. Band at 1200 bp is a product of RHCE gene while band 600 bp is the product from intron region of RHD gene. None of the five samples shown in the image carried intron 4 region of the RHD gene while both the bands were observed in the control sample.
RHD genotyping reaction with primer pair B1-B2 and C1-C2 is shown in Fig. 2, illustrating the pattern of reaction observed for exon 10 and 4 regions of RHD gene. None of the samples tested (0/200) showed the presence of RHDΨ gene. Sample detected as having intron 4 region upon testing with A1-A2 primer pair, showed presence of both, exon 10 and exon 4 products upon amplification with primer pair B1-B2 and C1-C2 respectively. Target segment in exon 4 region of RHD gene was not detected in any of the rest 199 samples tested. Exon 10 was detected in additional four samples (5 of total 200 samples tested). PCR reaction of sample identified as having all three regions tested in the study are shown in Fig. 3.
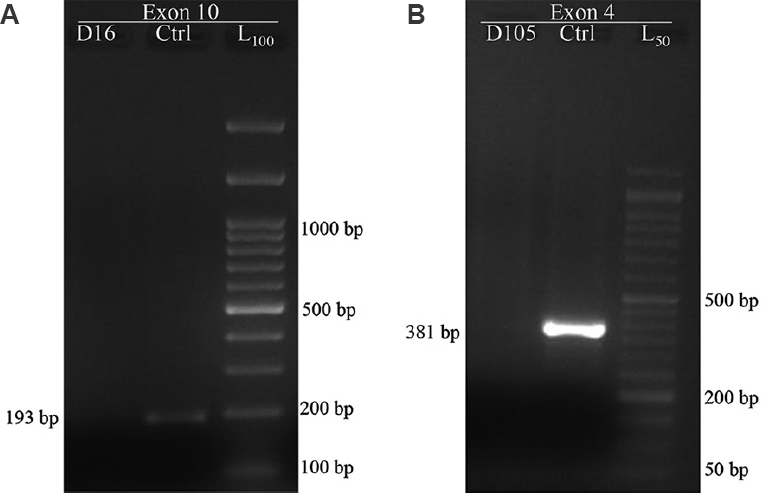
- PCR-sequence specific priming products for exon 10 region and normal / RHDΨ segment in exon 4 region of RHD gene (n=200). L100 - 100 bp DNA ladder, L50 - 50 bp DNA ladder. Reaction with primer set B1-B2. (A) shows a 193 bp product in the control sample (Ctrl) and no product in donor sample (D61). Reaction with primer set C1-C2. (B) shows a 381 bp product obtained in control sample and no product in donor (D105) sample. Presence of pseudo gene (RHDΨ) would have resulted in a 418 bp product instead of 381 bp product seen in sample with normal RHD gene.
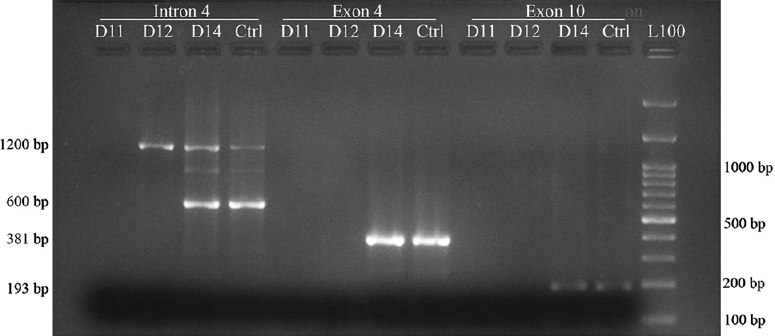
- PCR-sequence specific priming and gel electrophoresis for presence of RHD gene in an individual with RhD-negative phenotype. L100 - 100 bp DNA ladder, L50 - 50 bp DNA ladder. Reaction of study samples (D11, D12, D14) with all three primer sets used in the study are shown here. D14 sample amplification resulted in all three products expected from a normal RHD gene as in the control sample (Ctrl) from normal RhD-positive individuals. Only 1200 bp products were observed for sample D12 representing amplification of region in the homologous portion of RHCE gene.
Thus overall, no PCR product was detected in 195/200 (97.5%) of study samples indicating homozygous gene deletion. Of the 5/200 (2.5%) showing RHD gene polymorphisms, 4/200 (2%) were positive for the presence of exon 10 only (RHD-CE-D hybrid). RHDΨ gene was not detected in any of the samples tested. One sample showed presence of all three tested regions and was negative for RHDΨ gene. Details of samples detected with RHD gene polymorphism are shown in Table II.
| Extended Rh phenotype | Probable genotype | RHD gene status | Remarks | ||||||
|---|---|---|---|---|---|---|---|---|---|
| C | c | E | e | Intron 4 | Exon 10 | Exon 4 | RHDΨ | ||
| +4 | −ve | −ve | +3 | RHCe/RHCe | Present | Present | Present | Absent | Inactivating mutation in RHD gene? |
| −ve | +4 | −ve | +4 | RHce/RHce | Absent | Present | Absent | Absent | Presence of RHD-CE-D hybrid? |
| +4 | −ve | −ve | +3 | RHCe/RHCe | Absent | Present | Absent | Absent | |
| +4 | −ve | −ve | +3 | RHCe/RHCe | Absent | Present | Absent | Absent | |
| +4 | +4 | +1 | +4 | RHCE/RHce | Absent | Present | Absent | Absent | |
| RHCe/RHcE | |||||||||
Discussion
In the present study, the molecular basis of RhD-negative phenotype in Indian population was studied and as per our knowledge, so far, ours is the only study from Indian subcontinent on the prevalence of RHD gene in serologically RhD-negative individuals.
Inheritance of Rh antigens was explained by Tippett20, who proposed a two loci theory. According to this model, RH genes consist of two structural genes: one encoding the RhD antigen and the other encoding both the RhC/c and RhE/e antigens. Colin et al7 provided the evidence about existence of two highly homologous genes and that RhD-negative individuals carry only one gene. Later on, studies involving cloning of the RHD-cDNA, proved the absence of the RHD gene in RhD-negative individuals1721.
These two genes (RHD, RHCE), consisting of 10 exons each, are in close proximity on the short arm of chromosome one22 at location 1p34.1-1p36, encompassing 69 kbp of DNA. These genes encode both the Rh proteins (RhD and RhCE). One carries the D antigen and other carries CE antigens in various combinations (ce, Ce, cE, or CE). Individuals who lack RhD protein, ‘Rh or D negative’, most often have a complete deletion of the RHD gene. The difference between two highly homologous genes is on intron 4, where RHD contains a deletion of 600 bp in relation to RHCE23. This characteristic was utilized in the current study where a primer set was used on the sample of an RhD-positive individual to yield two products, one from the RHD gene (600 bp) and another from RHCE gene (1200 bp).
In African blood donors19, a 37 bp insertion in exon 4 (Rh pseudo gene) and other mutations have been reported to result in RhD-negative phenotype. Approximately 66 per cent of South African, D-negative, Black persons were reported to have RHD with a 37 bp internal duplication that causes a premature stop codon and does not encode a functional protein. Another mechanism for RhD-negative phenotype is presence of a hybrid gene (RHD-CE-D or RHCE-D-CE) resulting in deletion of certain portion of RHD gene and replacement with gene sequence from RHCE gene. All three probable causes were explored in the present study.
Thus, the three regions were included to identify the three most commonly reported reasons for RHD-negative phenotype in an individual despite the presence of the RHD gene. The highly conserved region of intron 4 was used to identify the presence of RHD gene as well as to ensure that the amplicon is not from the highly homologous RHCE gene. Region from exon 4 was chosen to identify the presence of any pseudogene and region from exon 10 that was used to identify the presence of RHD-CE hybrids formed as a result of crossover.
We found no PCR product in most (97.5%) of study samples indicating homozygous gene deletion and only 2.5 per cent of our study samples were found to have RHD gene polymorphisms. Our results are, however, in contrast to reports from other parts of the world. In a study from Japan8 on 130 RhD-negative donors, as many as 27.7 per cent demonstrated presence of RHD gene polymorphism by PCR. In a study from China24, a total of 204 RhD-negative blood donor samples were investigated by a modified PCR - restriction fragment length polymorphism (RFLP) and RT-PCR, the authors reported RHD gene deletion in 73.5 per cent of the cases and RHD-CE-D hybrid in 6.4 per cent of their samples. The authors of this study reported another mechanism Del, a deletion of 1013 bp between introns 8 and 9 including exon 9 of the RHD gene in 20.1 per cent of cases. Our findings are closer to the reports from Brazil25, with total RHD gene deletion in 95.8 per cent, while 4.1 per cent showed RHD gene polymorphisms.
In the present study, we observed that the sample with presence of all three portions of the RHD gene (serial 1 in Table II) belonged to an individual with RHCe/RHCe probable genotype. In the study from Japan8, the phenotypes of RhD-negative samples showing presence of RHD gene were CC or Cc, but not cc. It is therefore, suggested that there is some relationship between the RHD gene and the RhC phenotypes in RhD-negative individuals.
The findings of the current study are especially important in cases of RhD typing discrepancy and use of molecular testing methods for assessing the RHD type of foetus. This may prevent false or incorrect RHD typing results based on molecular testing as the foetus may actually be RHD-negative in spite of the presence of RHD gene.
It is therefore clear that ours was a fairly homogenous population with regard to molecular mechanism of RhD-negative haplotype as only 2.5 per cent of our study population was found to have RHD polymorphism. However, one area of concern may be incomplete coverage of the RHD gene in our study, as only three out of total 10 exons in RHD gene were screened and partial deletion of unscreened portion of RHD gene leading to RhD-negative phenotype might have been missed in the sample showing all three products.
Based on the results of the present study, it is recommend that testing of pseudo gene may be omitted from molecular typing of RHD status in India. RHD genotyping using a combination of two primer sets, one for intron 4 and one for exon 10 in Indian population may give false results in 2.5 per cent of the cases.
India is a country with huge ethnic diversity which is also reflected in the varying RhD phenotype status as reported from various parts of the country, ranging from 2 to ~ 8 per cent. Similar variation in genotype frequencies are also expected. Since the study involves North Indian population, the results may not be generalized and a multicentric study involving donors from across the country needs to be done.
According to the above-mentioned data, it is clear that the RHD gene is not as highly prevalent in our population as in other ethnic RhD-negative groups. A study from Switzerland26 suggested the use of an algorithm for assessing actual RhD status of apparently RhD-negative individuals considering the high incidence of occurrence of RHD gene in their RhD-negative donor population. However, based on our results, we do not recommend the same. RHD genotyping should be done only when there is a discrepancy in RhD grouping results or in special clinical conditions such as in the investigation of foetal RhD type before birth for making a rapid diagnosis and achieving a good prognosis in suspected cases of haemolytic disease of foetus and newborn.
Overall, although there are inherent limitations of serological testing, serologic typing should still be considered as the standard method to determine RhD phenotype and formulate transfusion strategies in Indian population.
Financial support & sponsorship: The present study received financial support as from an Intramural grant from Sanjay Gandhi Postgraduate Institute of Medical Sciences, Lucknow.
Conflicts of Interest: None.
References
- Applying molecular immunohematology discoveries to standards of practice in blood banks:Now is the time. Transfusion (48):2461-75.
- [Google Scholar]
- Integrating molecular technologies for red blood cell typing and compatibility testing into blood centers and transfusion services. Transfus Med Rev. 2008;22:117-32.
- [Google Scholar]
- The potential of blood group genotyping for transfusion medicine practice. Immunohematology (24):190-5.
- [Google Scholar]
- Large-scale blood group genotyping:Clinical implications. Br J Haematol. 2009;144:3-13.
- [Google Scholar]
- Molecular configuration of Rh D epitopes as defined by site-directed mutagenesis and expression of mutant Rh constructs in K562 erythroleukemia cells. Blood (94):3986-96.
- [Google Scholar]
- Variants of RhD–current testing and clinical consequences. Br J Haematol (161):461-70.
- [Google Scholar]
- Genetic basis of the RhD-positive and RhD-negative blood group polymorphism as determined by Southern analysis. Blood. 1991;78:2747-52.
- [Google Scholar]
- The RHD gene is highly detectable in RhD-negative Japanese donors. J Clin Invest. 1997;100:373-9.
- [Google Scholar]
- Rh genotyping:Avoiding false-negative and false-positive results among individuals of African ancestry. Am J Hematol. 2002;69:34-40.
- [Google Scholar]
- An easy RHD genotyping strategy for D- East Asian persons applied to Korean blood donors. Transfusion. 2006;46:2128-37.
- [Google Scholar]
- Potential of commercial anti-D reagents in the identification of partial D variants in Indian population. Indian J Med Res. 2007;125:641-4.
- [Google Scholar]
- Rh phenotype, allele and haplotype frequencies among 51,857 blood donors in North India. Blood Transfus. 2014;12:36-9.
- [Google Scholar]
- A simple diagnostic strategy for RhD typing in discrepant cases in the Indian population. Blood Transfus. 2013;11:37-42.
- [Google Scholar]
- RHD positive alleles among D-C/E+individuals from India. Transfus Med Hemother (45):173-7.
- [Google Scholar]
- Molecular cloning of RhD cDNA derived from a gene present in RhD-positive, but not RhD-negative individuals. Blood. 1993;82:651-5.
- [Google Scholar]
- Prenatal determination of fetal RhD type by DNA amplification. N Engl J Med. 1993;329:607-10.
- [Google Scholar]
- The presence of an RHD pseudogene containing a 37 base pair duplication and a nonsense mutation in africans with the Rh D-negative blood group phenotype. Blood. 2000;95:12-8.
- [Google Scholar]
- Molecular cloning and primary structure of the human blood group RhD polypeptide. Proc Natl Acad Sci U S A. 1992;89:10925-9.
- [Google Scholar]
- Evolution of the human RH (rhesus) blood group genes:A 50 year old prediction (partially) fulfilled. Hum Mol Genet. 1997;6:843-50.
- [Google Scholar]
- Review:The molecular basis of the Rh blood group phenotypes. Immunohematology. 2004;20:23-36.
- [Google Scholar]
- Molecular basis for the RhD negative phenotype in Chinese. Int J Mol Med. 2003;11:515-21.
- [Google Scholar]
- RHD allelic identification among D-Brazilian blood donors as a routine test using pools of DNA. J Clin Lab Anal (26):104-8.
- [Google Scholar]
- Molecular RHD screening of RhD negative donors can replace standard serological testing for RhD negative donors. Transfus Apher Sci. 2014;50:163-8.
- [Google Scholar]






