Translate this page into:
Kinetics of viral RNA, immunoglobulin-M & G antibodies in Kyasanur forest disease
For correspondence: Dr Devendra T. Mourya, ICMR-National Institute of Virology, 20-A, Dr. Ambedkar Road, Pune 411 001, Maharashtra, India e-mail: dtmourya@gmail.com
-
Received: ,
This is an open access journal, and articles are distributed under the terms of the Creative Commons Attribution-NonCommercial-ShareAlike 4.0 License, which allows others to remix, tweak, and build upon the work non-commercially, as long as appropriate credit is given and the new creations are licensed under the identical terms.
This article was originally published by Wolters Kluwer - Medknow and was migrated to Scientific Scholar after the change of Publisher.
Abstract
Background & objectives:
Kyasanur forest disease (KFD) is an infectious disease discovered in Karnataka State of India in 1957; since then, the State has been known to be enzootic for KFD. In the last few years, its presence was observed in the adjoining five States of the Western Ghats of India. The present study was conducted to understand the kinetics of viral RNA, immunoglobulin M (IgM) and IgG antibody in KFD-infected humans for developing a diagnostic algorithm for KFD.
Methods:
A prospective follow up study was performed among KFD patients in Sindhudurg district of Maharashtra State, India. A total of 1046 suspected patients were tested, and 72 KFD patients were enrolled and followed for 17 months (January 2016 to May 2017). Serum samples of KFD patients were screened for viral RNA, and IgM and IgG antibodies.
Results:
KFD viral positivity was observed from 1st to 18th post-onset day (POD). Positivity of anti-KFD virus (KFDV) IgM antibodies was detected from 4th till 122nd POD and anti-KFDV IgG antibodies detected from 5th till 474th POD. A prediction probability was determined from statistical analysis using the generalized additive model in R-software to support the laboratory findings regarding viral kinetics.
Interpretation & conclusions:
This study demonstrated the presence of KFD viral RNA till 18th POD, IgM antibodies till 122nd POD and IgG till the last sample collected. Based on our study an algorithm was recommended for accurate laboratory diagnosis of KFDV infection. A sample collected between 1 and 3 POD can be tested using KFDV real-time reverse transcriptase polymerase chain reaction (RT-PCR); between 4 and 24 POD, the combination of real-time RT-PCR and anti-KFDV IgM enzyme-linked immunosorbent assay (ELISA) tests can be used; between POD 25 and 132, anti-KFDV IgM and IgG ELISA are recommended.
Keywords
Diagnostic algorithm
human
IgM/IgG antibody
Kyasanur forest disease
Maharashtra
viral RNA
Kyasanur forest disease virus (KFDV), belonging to the family Flaviviridae, is considered as one of the high-risk category pathogens. This virus is regarded as highly infectious, as shown by various field and laboratory studies1. Until 1971, this disease was enzootic in Shimoga district of Karnataka State, but recent reports revealed the broadened KFD arena in the new realm of Karnataka, Kerala, Tamil Nadu, Goa and Maharashtra2345.
Earlier researchers have demonstrated that the viraemia remains high during the first 3-6 days attaining titre 3.1×106 plaque-forming unit (pfu)/ml, and it remains high in some cases up to 10-12 days after onset of symptoms6. Clinical outcomes include sudden chills, acute fever, headache and myalgia and often exhibit biphasic nature of the illness. Limited studies have been conducted to understand the kinetics of viraemia in KFD-infected individuals78. Shah et al9 also conducted challenge studies in monkeys and mice. These studies were based on neutralization assays performed on monkey serum samples and were used to detect the level and duration of persisting viraemia in monkeys. Further, histopathological studies were conducted to study the degree of tissue damage occurring in the infected organs of monkeys10.
The present study was conducted on a prospective group of 72 individuals who were diagnosed positive for KFDV infection. Parameters analyzed in the study included viral copy numbers along with immunoglobulin M (IgM) and IgG profiles generated during the infection to develop algorithm for laboratory diagnosis of KFDV in infection. Data generated from these analyses would help in improved evaluation for the diagnostic period about the KFDV and early detection of KFDV infection. It would also help in designing novel vaccines or for drug trials to be conducted in future studies.
Material & Methods
Institutional Human Ethics Committee (IHEC) at the Indian Council of Medical Research (ICMR)-National Institute of Virology (NIV), Pune, approved the study conducted (NIV/IHEC/2015/D-5852). Informed written consents were obtained from all the study participants and parent/guardian of the children included in this study.
Study site and inclusion criteria: KFDV-affected areas in Dodamarg and Sawantwadi Taluka of Sindhudurg district of Maharashtra State were selected as the study site from January 2016 to May 2017. After obtaining written informed consent from the patients suspected to have KFD11 and who fulfilled the inclusion criteria, 3 ml blood samples were collected from each patient attending OPD at Rural Hospital Dodamarg, Sindhudurg, Maharashtra in two tubes containing Ethylenediaminetetraacetic acid (EDTA) and clot activator. The inclusion and exclusion criteria for the sample included and analyzed were (i) viral RNA-positive status and/or the presence of IgM antibodies for KFDV, and (ii) date of onset between January 2016 and May 2017 (Fig. 1A & B).
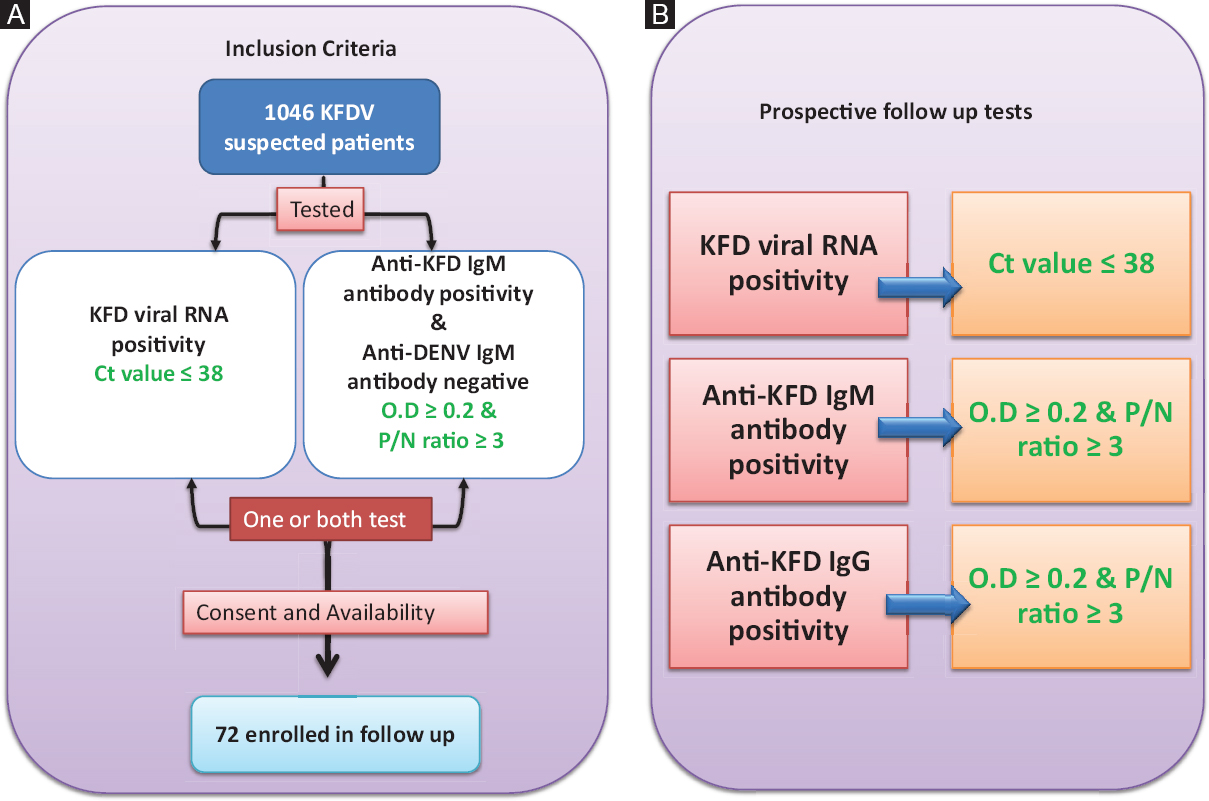
- Overview of the study design: (A) Inclusion criteria used to select follow up patients. (B) Prospective follow up analysis of 72 patients in the study. Green colour texts are the outcome of the study. KFDV, Kyasanur forest disease virus; Ig, immunoglobulin; Ct, cyclic threshold; OD, optical density; P/N, Positive/Negative ratio.
Collection of the samples from KFD patients and patient characteristics: Three hundred eighty three human serum samples were collected from 72 KFD patients at different time points spanning 474 days (17 months). Of the 72 patients, two patients gave sample eight times, seven patients gave sample seven times, 22 patients gave sample six times, 29 patients gave sample five times, seven patients gave sample four times and three patients gave sample three times while two patients gave sample two times. Blood samples were transported from Rural Hospital, Dodamarg, Sindhudurg in the cold chain to ICMR-NIV, Pune. Sample from each patient was registered, and a unique identification number was allotted to maintain the secrecy of the patient's identity.
Diagnostic parameters such as cyclic threshold value (Ct value) for real-time reverse transcriptase polymerase chain reaction (RT-PCR), optical density (OD) and positive to negative (P/N) ratio in the case of anti-KFDV IgM enzyme-linked immunosorbent assay (ELISA) and anti-KFDV IgG ELISA assays were generated (Fig. 1B). A clinical-epidemiological study was performed on identifying KFD patients as described in previous work12.
Detection of viral RNA, IgM and IgG antibodies to understand its persistence in KFD patients: All the samples were processed with appropriate bio-safety precautions; the serum samples were inactivated in a containment laboratory and used for RNA extraction and ELISA testing in the Biosafety Level 2 (BSL-2) facility. Serum was separated from the collected blood sample and was used for extracting RNA and serological analysis as described earlier13. RNA extracted from each sample was diagnosed using real-time RT-PCR and anti-KFDV IgM ELISA and anti-KFDV IgG ELISA assays as described previously13. The presence of viral RNA in the serum was determined using real-time RT-PCR and defined to be positive if it had a Ct value of ≤3813 (Fig. 1B). A modified anti-KFDV IgM capture ELISA was used where the KFD mouse brain-derived antigen was replaced with tissue culture-grown KFD antigen. Further to increase the sensitivity, biotinylated antibodies and streptavidin-horseradish peroxidase (HRP) were used as detection antibodies. A sample was considered to be positive for anti-KFDV IgM antibodies if the OD of a sample was ≥0.2 and P/N ratio was ≥3, or else the sample was considered negative (Fig. 1B). ELISA plates (Nunc, Thermo Fisher Scientific, USA) were coated with gamma-inactivated KFDV antigen (Row A-D) for anti-KFDV IgG antibody and BHK-21 cells were used as control antigen (Row E-H) in carbonate buffer (pH 9.6, 0.05 M) overnight at 4°C. Subsequently, wells were blocked using liquid plate sealer (Candor Biosciences, Germany). The wells were vacuum-dried, sealed, and stored at 4°C until further use. One hundred microlitres (μl) of diluted sample (1:600 in 1% BSA in 1x PBS containing 0.1% tween-20) was added to duplicate wells of KFDV antigen and control antigen-coated wells and incubated at 37°C for one hour. One hundred microlitres of anti-human IgG HRP antibodies (1:30,000) (Pierce, USA) were added and further incubated for one hour at 37°C. The wells were washed four times using 10 mM PBS, pH 7.2 and 0.1 per cent Tween-20 (Sigma, USA) at the end of each step. One hundred microlitres of 3,3',5,5'-Tetramethylbenzidine (TMB) substrate (Clinical Biosciences, USA) was added and incubated for 10 min. The reaction was stopped using 1N H2SO4 and the absorbance was measured at 450 nm. The sample was considered positive for anti-KFDV IgG antibodies if the OD of the sample was ≥0.2 and P/N ratio was ≥3, or else the sample was considered as negative (Fig. 1B).
Both the ELISA assays were checked for their specificity using known positive and negative controls against other flaviviruses such as Japanese encephalitis, West Nile, and dengue virus (DENV). Twenty per cent of cross-reactivity was observed with DENV IgM and IgG-positive controls, which is a common observation across flaviviruses14. As per the inclusion criteria, only KFDV-positive cases were analyzed, avoiding even the lowest probability of false-positive status with DENV. All the KFDV IgM antibody-positive samples were also tested for dengue IgM antibodies to rule out this possibility and were found to be negative.
All the samples obtained were either negative or positive for a given test and were grouped into bins of the post-onset day (POD) for the per cent positivity analysis. RNA positivity in a sample was observed during the initial POD and had a narrow range; hence, the bins created were smaller in the initial phase to understand the dynamics of the KFD RNA positivity in the follow up study. The size of the bins was broadened for the anti-KFDV IgM detection and anti-KFDV IgG detection analysis as these had an extended detection period (Table).
| POD group | Per cent positive | ||
|---|---|---|---|
| KFD RNA-positive/patient tested | IgM-positive/patient tested | IgG-positive/patient tested | |
| 0-4 | 24/24 (100) | 1/24 (4.17) | 0/24 (0.00) |
| 5-10 | 15/29 (51.72) | 17/29 (58.62) | 11/29 (37.93) |
| 11-20 | 4/29 (13.79) | 27/29 (93.10) | 18/29 (62.68) |
| 21-30 | 0/22 (0.00) | 19/22 (86.36) | 18/22 (81.82) |
| 31-40 | 0/26 (0.00) | 22/26 (84.62) | 24/26 (92.31) |
| 41-60 | 0/32 (0.00) | 22/32 (68.75) | 32/32 (100.00) |
| 61-143 | 0/37 (0.00) | 15/37 (40.54) | 37/37 (100.00) |
| 170-210 | 0/46 (0.00) | 0/46 (0.00) | 40/46 (86.96) |
| 211-303 | 0/32 (0.00) | 0/32 (0.00) | 30/32 (93.75) |
| 313-381 | 0/45 (0.00) | 0/45 (0.00) | 27/45 (60.00) |
| 382-400 | 0/22 (0.00) | 0/22 (0.00) | 12/22 (59.52) |
| 401-440 | 0/24 (0.00) | 0/24 (0.00) | 13/24 (54.17) |
| 441-480 | 0/17 (0.00) | 0/17 (0.00) | 10/17 (58.82) |
Values in parentheses represent percentages. POD, post-onset day
Statistical analysis: The data for 72 patients had the limitations of uneven sample points and time points from each patient. Thus, to reduce the influence of the missing data points, the statistical analysis was used to extrapolate these existing data for predicting the rare possibility to estimate the different outcome variables of this study.
The KFD viral copy numbers and OD values along with the P/N ratio for anti-KFDV IgM and IgG antibodies were analyzed using generalized additive models (GAMs) in R-software version 3.4.3 for windows1516. The analysis was carried out on the basis of a smooth spline generated from POD. Two different kinds of univariate models were formed: (i) Gaussian error distribution using identity as the linked function, and (ii) binomial error distribution using logit as the linked function. Different programmes were written to obtain the statistical output.
Results
The study included 72 patients belonging to the age group of 2-75 yr with mean age being 19.5 yr having an intraquartile range between 28 and 55 yr, of which 65.2 per cent were female.
Detection of viral RNA, IgM and IgG antibody to understand its persistence in KFD patients: Of the 72 patients, 34 were KFDV RNA positive; 30 were anti-KFDV IgM positive while eight patients were both KFD viral RNA and anti-KFDV IgM antibody positive. For graphical representation of the data (Fig. 2), samples were grouped into various POD ranges as given in the Table so as to include a minimum of 15 samples in one group. Per cent positivity was calculated for each group and graph was plotted. Blue, red and green lines of Fig. 2 depict the per cent positive status of the KFD RNA, anti-KFDV IgM detection and anti-KFDV IgG detection, respectively, by clustering data at different POD ranges (Table). It was observed that there was an approximately 100 per cent occurrence of KFD RNA positivity detection during the initial four PODs which consequently reduced (51.72; 13.79%), and above 21 POD, there was no KFDV RNA positivity observed. The detection of anti-KFDV IgM detection commenced from the fourth POD and increased gradually following the reduction in detection peak of real-time RT-PCR. A maximum per cent positivity for anti-KFDV IgM detection of 93.10 per cent was observed during the 11th-20th POD. This detection gradually decreased after 20th POD till 122nd POD beyond which positivity was not recorded. Anti-KFDV IgG antibody positivity was observed after the fifth POD and increased drastically till 60th POD. The antibody remained detectable till the last time-point of sample collection (474 POD); however, a per cent positivity of 100 per cent was observed during 41st-140th POD range and then decreased gradually. The Table gives the detail of the positivity found for KFD diagnostic markers at different bins of the POD.
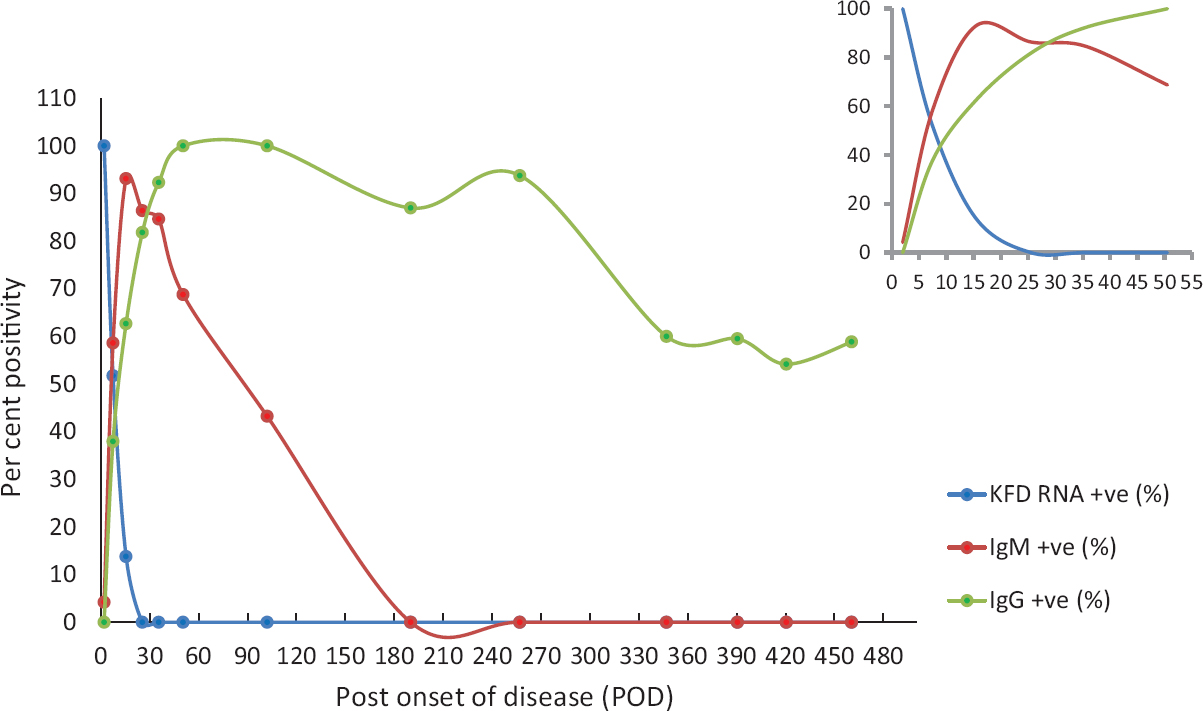
- Response in terms of per cent positivity for diagnostic markers at varying post onset of disease (POD). Blue, red and green lines depict the per cent positivity response for real-time RT-PCR, anti-KFDV IgM antibody, and anti-KFDV IgG antibody, respectively. The x-axis is increasing values of days post-onset of illness and y-axis depicts the fitted values of the observed response. Inset determines the per cent positivity during the initial 40 POD.
Figure 3A depicts observed viral RNA copy number on symptom onset to sample collection (POD) using R-software. The mean KFDV copy number was obtained to be 2.4×106 with the intraquartile range of 1.1×105 to 2.5×106. Viral RNA copy number was observed to be elevated from the 3rd to 6th POD. The mean viral RNA copy number was comparable between male and female as depicted in Fig. 3B. Figure 3C–F show the relative amount of OD data and P/N ratio observed for IgM antibody and IgG antibody, respectively, during each POD for 383 samples. The observed values were used to determine the positive status of the suspected samples obtained from patients.
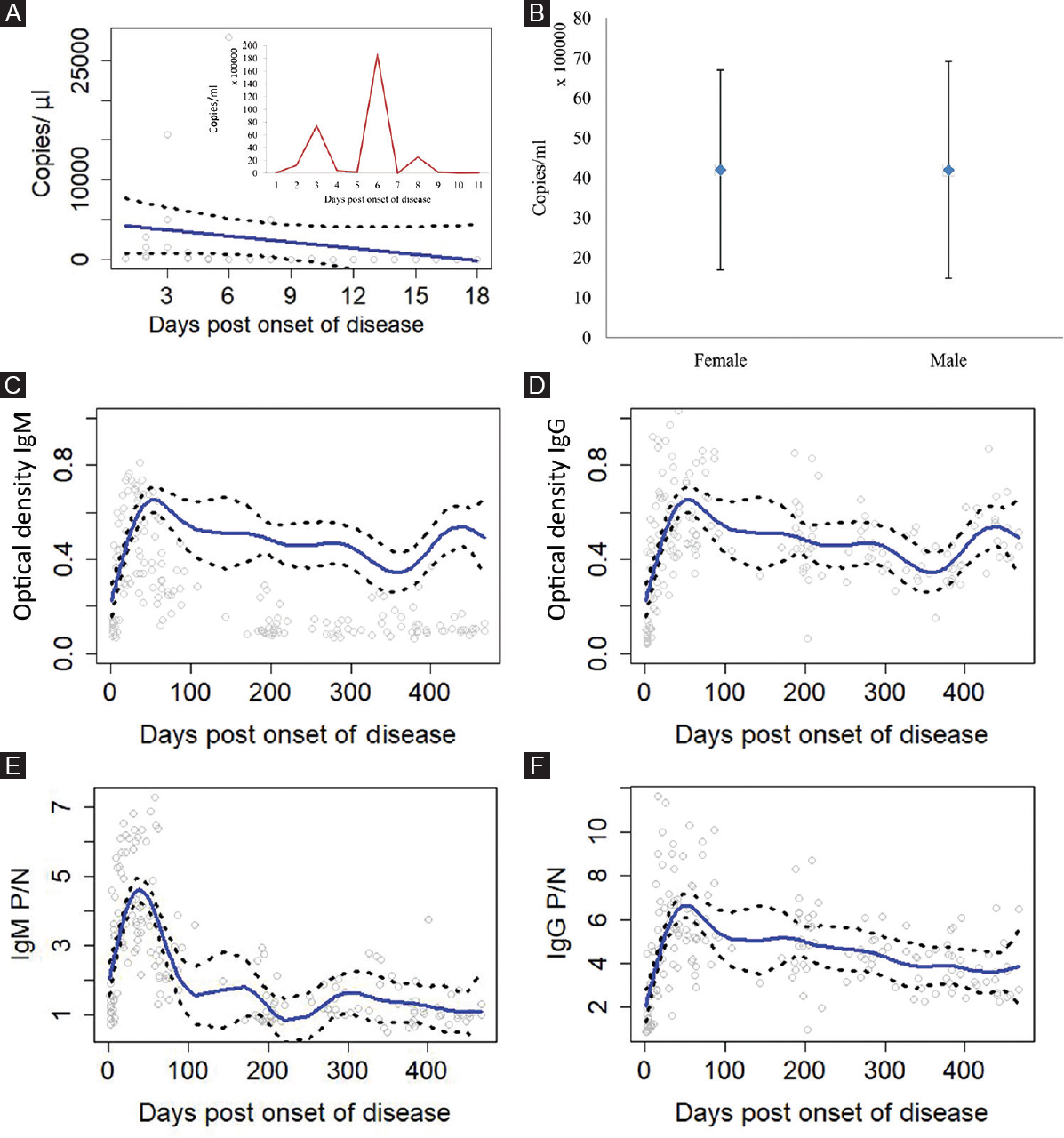
- (A) Relative amounts of Kyasanur forest diseases virus (KFDV) copy number observed at the different post onset of diseases (POD). Inset figure shows the average KFDV copy number at a particular POD. (B) Gender-wise distribution of mean KFDV copy number, (C and D) Optical density values observed for IgM and IgG antibodies at the different POD for 383 samples. (E and F) P/N ratio observed for IgM and IgG antibodies at the different POD for 383 samples. Solid lines are mean predicted values and dashed lines represent values enclosing 95 per cent of the mean. P/N, positive to negative.
Viral RNA kinetics, IgM and IgG antibody persistence in KFD patients using R-software: A probability value for the samples to be detected positive or negative was obtained using the GAM in R-software. KFDV RNA was detected earliest by the first POD with a probability of a sample to be 92.8 per cent positive [99.8-85.9%: 95% confidence interval (CI)]. Positivity decreased till 23rd POD, where there was one per cent chance for a sample to be real-time RT-PCR positive (17-29 POD: 95% CI). Anti-KFDV IgM antibodies had 29 per cent probability to be detected positive from the fourth POD. The IgM antibody positivity increased to 90 per cent on the 22nd POD (917-31 POD: 95% CI). The least (1%) chance for a sample to be detected positive for anti-KFDV IgM antibody was on the 140th POD (130-149 POD: 95% CI). Secondary antibody response, i.e. anti-KFDV IgG antibody had a detection probability of 22 per cent on the fifth POD and 27 per cent on the seventh POD. Its detection increased to 90 per cent on the 31st POD (25-37 POD: 95% CI). Anti-KFDV IgG antibody was predicted with a probability of being positive 62 per cent till the last observed on the 474th POD.
Discussion
Due to spread of KFD into newer areas, the data associated with the KFD infections are limited. Viral RNA of flaviviruses has a highly variable range of detection after infection. Paz-Bailey et al17 detected Zika virus RNA in serum sample as late as 54 days, while Hirayama et al18 could detect DENV in the serum till the 11th day. KFD often causes biphasic fever; therefore, in the present study, 383 blood samples from 72 patients until 474 days were analyzed. It was necessary to have data from a longitudinal survey of KFD for designing a proper algorithm for KFD diagnosis. Early, sensitive and specific diagnosis can be made when such an algorithm is applied during outbreak investigations. It can also assist in understanding the time point for sample collection during epidemiological and cross-reactivity studies.
With the present study, the KFD viral RNA positivity was 100 per cent between the 1st and 4th POD, and this decreased beyond the fifth POD, the last sample being positive for KFD viral RNA on the 18th POD. Immune response mounted was detected using IgM and IgG antibodies against KFDV. Primary immune response mounted in the form of IgM antibodies started appearing only after the fourth POD. Secondary immune responses observed in terms of IgG were detected after the fifth POD, and it persistently increased.
The prediction from the current data suggested that KFDV copy number in the serum could be detected up to the 24th POD with one per cent probability. The antibody response in the form of IgM was detected with 92 per cent probability of IgM to be positive on the 24th POD, which lasted till the 132nd POD in one per cent of samples. The prediction possibility for IgG was 79 per cent from the 25th to 98 per cent on 132nd POD, which further decreased to 61.5 per cent on the 474th POD (Fig. 4).

- Algorithm for accurate KFD laboratory diagnosis: The figure shows the recommend test accordance with the POD for KFD-affected patients. Rectangles depicts the POD range and numbers in the parenthesis indicate the probability of percentage positivity. KFDV, Kyasanur forest disease virus; POD, post-onset day; ELISA, enzyme-linked immunosorbent assay; RT-PCR, Reverse transcriptase polymerase chain reaction.
This study provided information on the overall situation on the kinetics of viral RNA, along with the diagnostic markers for the immune response (IgG and IgM) in the KFD patients. Using the observed and predicted kinetics values, a combination of test is proposed that should be performed for confirmation of KFD infection in the patients to have maximum accuracy in the diagnosis. Based on the data, an algorithm was designed for efficient and timely diagnosis of KFDV infection (Fig. 4).
Initially, a systematic periodic collection of defined periods (days, weeks and months) was proposed to monitor the progress of viral RNA, IgM and IgG antibody in patients. However, due to the non-availability of many patients for follow up periods or non-consent for follow up proposed periodic sample collection, the data were not available for the analysis. Fewer sample points during the POD range of 80-120 was the major limitation of the current study.
To conclude based on the present data, an algorithm was proposed for the efficient and timely diagnosis of KFD infection. If the sample was collected at POD between 1 and 3, KFDV real-time RT-PCR was recommended; between 4 and 24 POD, the combination of real-time RT-PCR and anti-KFDV IgM ELISA tests would be a test of choice; between POD 25 and 132, anti-KFDV IgM and IgG ELISA would be preferred; beyond 132nd POD, it would be advisable to perform only anti-KFDV IgG ELISA assay.
Acknowledgment
Authors acknowledge the contribution of Sarong GD and Andhare MD [Medical Officer, Primary Health Centers (Sateli-Bhedshi and Morgaon), Taluka Dodamarg, District Sindhudurg], Sale YR (District Health Officer, Health Department, Zilla Parishad, Oras, District Sindhudurg) of district surveillance unit, Maharashtra State, for collection of clinical samples of suspected cases and necessary clinical data. Authors also acknowledge the contribution of technical staff from BSL-4 laboratory, NIV, Pune, Shrimati Shital Dalal and Shrimati Yogita Chopade, Servshri Pravin Kore, Sanjay Gopale, Kumar Bagmare, and Shrimati Amita Bargat.
Financial support & sponsorship: Financial support was provided by the ICMR-NIV, Pune, India.
Conflicts of Interest: None.
References
- Kyasanur forest diseases. In: Monath TP, ed. Arboviruses: Epidemiology and ecology. Boca Raton (FL): CRC Press; 1990. p. :93-116.
- [Google Scholar]
- Outbreak of Kyasanur forest disease (monkey fever) in Sindhudurg, Maharashtra State, India, 2016. J Infect. 2016;72:759-61.
- [Google Scholar]
- On the transmission pattern of Kyasanur forest disease (KFD) in India. Infect Dis Poverty. 2015;4:37.
- [Google Scholar]
- New focus of Kyasanur forest disease virus activity in a tribal area in Kerala, India, 2014. Infect Dis Poverty. 2015;4:12.
- [Google Scholar]
- Outbreak of Kyasanur forest disease in Thirthahalli, Karnataka, India, 2014. Int J Infect Dis. 2014;26:132-4.
- [Google Scholar]
- Kyasanur forest diseases. IV. Isolation of Kyasanur forest disease virus from infected humans and monkeys of Shimoga district, Mysore State. Indian J Med Sci. 1966;20:316-20.
- [Google Scholar]
- Clinical, clinicopathologic, and hematologic features of Kyasanur forest disease. Rev Infect Dis. 1989;11(Suppl 4):S854-9.
- [Google Scholar]
- Kyasanur forest disease in the human population of Shimoga district, Mysore State, 1959-1966. Indian J Med Res. 1975;63:1556-63.
- [Google Scholar]
- Kyasanur forest disease virus: Viremia and challenge studies in monkeys with evidence of cross-protection by Langat virus infection. F1000Res. 2012;1:61.
- [Google Scholar]
- Kyasanur Forest Disease. Part VII. Pathological findings in monkeys, Presbytis entellus and Macaca radiata, found dead in the forest. Indian J Med Res. 1960;48:276-86.
- [Google Scholar]
- CD Alert: Kyasanur Forest Disease: A Public Health Concern. Delhi: Directorate General of Health Services; Available from: https://idsp.nic.in/WriteReadData/l892s/60398414361527247979.pdf
- Kyasanur forest disease prevalence in Western Ghats proven and confirmed by recent outbreak in Maharashtra, India, 2016. Vector Borne Zoonotic Dis. 2018;18:164-72.
- [Google Scholar]
- Diagnosis of Kyasanur forest disease by nested RT-PCR, real-time RT-PCR and IgM capture ELISA. J Virol Methods. 2012;186:49-54.
- [Google Scholar]
- R: A language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing; 2017.
- Persistence of Zika virus in body fluids – Final report. N Engl J Med. 2018;379:1234-43.
- [Google Scholar]
- Detection of dengue virus genome in urine by real-time reverse transcriptase PCR: A laboratory diagnostic method useful after disappearance of the genome in serum. J Clin Microbiol. 2012;50:2047-52.
- [Google Scholar]






