Translate this page into:
Interleukin-1β (IL-1β) & IL-4 gene polymorphisms in patients with systemic lupus erythematosus (SLE) & their association with susceptibility to SLE
Reprint requests: Dr Saeedeh Salimi, Department of Clinical Biochemistry, School of Medicine, Zahedan University of Medical Sciences, Zahedan, Iran e-mail: sasalimi@yahoo.com
-
Received: ,
This is an open access article distributed under the terms of the Creative Commons Attribution-NonCommercial-ShareAlike 3.0 License, which allows others to remix, tweak, and build upon the work non-commercially, as long as the author is credited and the new creations are licensed under the identical terms.
This article was originally published by Medknow Publications & Media Pvt Ltd and was migrated to Scientific Scholar after the change of Publisher.
Abstract
Background & objectives:
Interleukin-1 (IL-1) is one of the pro-inflammatory cytokines that plays a main role in the regulation of immune and inflammatory responses. Interleukin 4 (IL-4) as an anti-inflammatory cytokine regulates balance between Th1 and Th2 immune responses. This study was undertaken to investigate the IL-1β and IL-4 genes polymorphisms in patients with systemic lupus erythematosus (SLE) and also association between the polymorphisms and susceptibility to SLE.
Methods:
One hundred and sixty three SLE patients and 180 healthy controls were genotyped for the IL-4 VNTR (variable number tandem repeat), IL-1β C-511T and IL-1β T-31C polymorphisms by polymerase chain reaction (PCR) or PCR-RFLP (restriction fragment length polymorphism) method.
Results:
The frequencies of CC genotype and C allele of the IL-1β T-31C polymorphism were significantly (P<0.01) lower in SLE patients than controls. Moreover, the frequencies of RP1/RP2 genotype and RP2 allele of IL-4 VNTR polymorphism were significantly (P<0.05) higher in the SLE patients. No association was observed between IL-1β C-511T polymorphism and increased risk of SLE. We observed increased frequency of CT and TT genotypes of IL-1β C-511T polymorphism in SLE patients with malar rash compared to SLE patients without this manifestation.
Interpretation & conclusions:
The present findings suggest that IL-1β T-31C and IL-4 VNTR polymorphisms but not IL-1β C-511T polymorphism may contribute in SLE pathogenesis. In addition, CT and TT genotypes of IL-1β C-511T polymorphism were associated with SLE.
Keywords
Interleukin-1β
interleukin-4
polymorphism
systemic lupus erythematosus
Systemic lupus erythematosus (SLE) is a heterogeneous autoimmune disease characterized by pathogenic autoantibody formation against the host's nuclear antigens, immune complex deposition, and end-organ damage1. The exact aetiology of this disease is unknown but genetic and environmental factors have important roles in SLE pathogenesis2. The prevalence of this disease in women is about 8-10 times more than men and in African Americans is about 3-4 times more than Caucasian Americans3. The prevalence of SLE has been reported to be 190 per 100,000 persons in South East Iran4. Family and genetic studies, especially investigations on monozygotic and dizygotic twins, indicate an essential role for genetic predisposition to SLE5. One of the potential susceptible regions for SLE is located in the human leukocyte antigen (HLA) region. However, this is not the only susceptible region in relation with SLE pathology6.
Given the importance of interleukins (ILs) in immune system regulation, several immunological abnormalities such as dysregulation of B-cell activation that leads to B-cell hyperactivity and overproduction of autoantibodies have been reported in SLE7. Several reports have indicated the correlation between SLE and different pro- inflammatory and anti- inflammatory interleukins including IL-1β and IL-489.
IL-1β, a member of IL-1 gene family (IL-1α, IL-1β and IL-1Ra) is a potential pro-inflammatory cytokine that plays an important role in inflammatory and immune responses. This cytokine is produced mainly by activated macrophages and monocytes10. However, the human IL-4 is an anti-inflammatory cytokine produced by CD4+ Th2 cells, basophils and mast cells which are involved in the regulation of the humoral immune response11. IL-4 has cytotoxic effect, inhibits induction of nitric oxide synthase and release of superoxide anions by macrophages, and has several anti-inflammatory effects. IL-1β and IL-4 genes are mapped on chromosomes 2q14-21 and 5q31-33, respectively1112. IL-1β gene contains two polymorphisms in its promoter region, including IL-1β T31C, IL-1β C-511T that cause higher expression of IL-1β. Considering the essential role of IL-1 in inflammation, it seems that allelic polymorphisms in IL-1 might have an effect on susceptibility to SLE. Moreover, there is a 70bp VNTR (variable number tandem repeat) polymorphism located in the third intron of IL-4 gene, which could be a candidate region for this disease. Several studies have investigated the association between IL-1β T31C, IL-1β C-511T and IL-4 70bp VNTR polymorphism and SLE susceptibility, however, the results were inconsistent913.
Given the involvement of IL-1β and IL-4 in autoimmune diseases including SLE89, the aim of this study was to investigate IL-1β, IL-4 genes polymorphism in patients with SLE and their possible association with susceptibility to SLE.
Material & Methods
This study was conducted on 163 consecutive SLE patients (13 men and 150 women) who were referred to the Rheumatology clinic of Ali-Ebne-Abitaleb hospital, Zahedan University of Medical Sciences, Zahedan, Iran, from June 2011 to May 2013. Individuals with other rheumatic diseases, infections or malignant tumours were excluded. All patients fulfilled at least four items of SLE, according to American College of Rheumatology (ACR) 1997 criteria14. A written informed consent was obtained from all participants, and the study protocol was approved by the Ethics Committee of Zahedan University of Medical Sciences, Zahedan, Iran.
The control group consisted of 180 age, sex and ethnically matched volunteers (14 men and 166 women) with negative antinuclear antibody (ANA) test and were randomly selected from healthy volunteers who were referred to the internal medicine clinic for general examination. The controls had no systemic disease and were not related to lupus patients.
Blood samples (2 ml) of all participants were collected in sterile EDTA tubes and kept frozen at -20°C.
Genomic DNA extraction and genotyping: Genomic DNA was extracted from peripheral blood leukocytes by salting out method15. Genotypes of IL-1β polymorphisms were detected using polymerase chain reaction-restriction fragment length polymorphism (PCR-RFLP) method. PCR amplification was performed for 70bp VNTR polymorphism of the IL-4 gene. The sequence of primers were as follows: for IL-1β C-511T polymorphism, forward 5’-TGGCATTGATCTGGTTCATC-3’ and reverse 5’-GTTTAGGAATCTTCCCACTT-3’16; for IL-1βT-31C polymorphism, forward 5’-AGAAGCTTCCACCAATACTC-3’ and reverse 5’-ACCACCTAGTTGTAAGGA-3’17; for IL-4 70bp VNTR polymorphism, forward 5’-AGGCTGAAAGGGGGAAAGC-3’ and reverse 5’-CTGTTCACCTCAACTGCTCC-3’18.
Polymerase chain reaction was performed in a 25 μl final volume containing 25 pmol of each primers, 0.1mM dNTP (Fermentas, Lithuania), 0.5 μg genomic DNA, 1.5 mM MgCl2, 2.5 μl of 10× PCR buffer and 1.5 U Taq DNA polymerase (Fermentas, Lithuania), according to the following protocol: initial denaturation at 94°C for four min; 30 cycles of denaturation at 94°C for 45 sec, annealing for 30 sec at 57°C for IL-1β C-511T and 61°C for IL-1β T-31C and IL-4 VNTR polymorphisms, and extension at 72°C for 45 sec; and final extension at 72°C for five min. The 305 bp PCR fragments of IL-1β C-511T polymorphism were digested with Ava I restriction enzyme (Fermentas, Lithuania) for 16 h at 37°C. The C allele had one Ava I cleavage site and digested to 115 and 190 bp fragments, whereas the T allele had no Ava I cleavage site and produced 305 bp fragment only.
The 239bp PCR product of IL-1β T-31C polymorphism was digested with Alu I restriction enzyme (Fermentas, Lithuania) for 16 h at 37°C. The T allele had Alu I cleavage site and digested to 152 and 87bp fragments, whereas the C allele had no Alu I cleavage site and produced 239bp fragment only. The PCR product of IL-4 70bp VNTR polymorphism was a 183bp fragment in the absence of repeat polymorphism1 (RP1) and a 253bp fragment in the presence of RP2 of the 70bp VNTR fragment. PCR and digested products were separated by electrophoresis on a 2.5 per cent agarose gel and visualized by ethidium bromide staining.
Statistical analysis: Data were analyzed using the statistical software SPSS 18 (SPSS, Chicago, IL, USA). The differences between groups were analyzed by independent Student t test, chi square test or Fisher's exact test, wherever appropriate. The genotypes and alleles frequency were compared between SLE patients and controls by χ2 test and Fisher's exact test. The odds ratio (OR) and 95% confidence intervals (CI) were also estimated. Logistic regression analysis was used to assess the independent effect of each risk polymorphism on SLE. Bonferroni's correction was applied to confirm the association of haplotypes with the disease. Linkage disequilibrium (LD) was analyzed using CubeX software19.
Results
Demographic data of SLE patients and control group are shown in Table I. There were no significant differences for age, gender and ethnicity between SLE patients and controls.
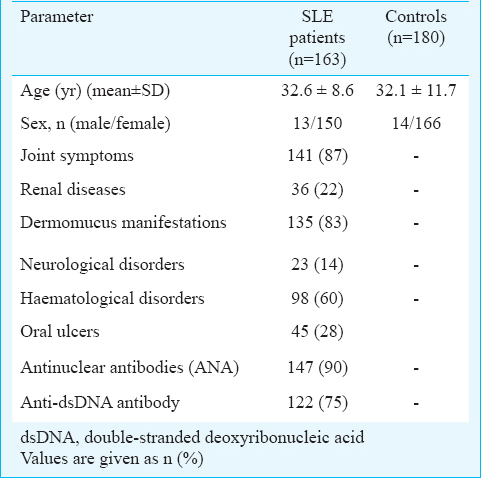
The allele and genotype frequencies of IL-1β C-511T, IL-1β T-31C and IL-4 VNTR polymorphisms in patients with SLE and controls are shown in Table II. All loci were in Hardy-Weinberg equilibrium. Linkage analysis showed that IL-1β promoter single nucleotide polymorphisms (SNPs) rs1143627 and rs16944 were located in one haplotype block and the magnitude of LD between the SNPs was high (D′=0.665, r2=0.324).
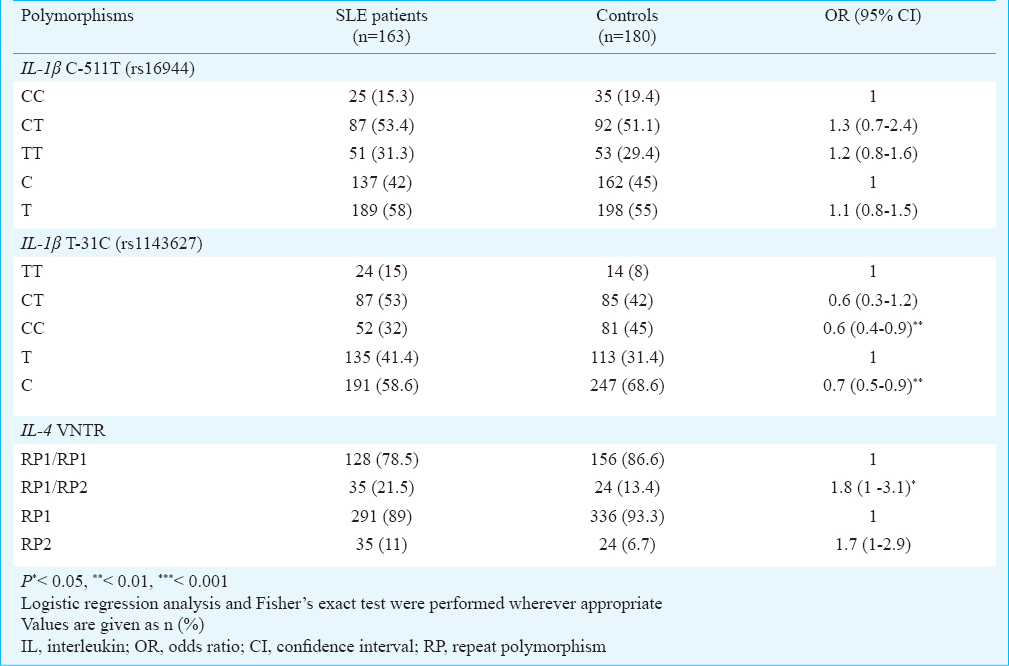
The genotype and allele frequencies of IL-1β C-511T polymorphism were not significantly different between SLE patients and control group. However, the genotype and allele frequencies of IL-1β T-31C polymorphism were significantly different between the two groups and the risk of SLE was lower in individuals with CC genotype compared to those with TT genotype. [OR, 0.6 (95% CI, 0.4 to 0.9); P<0.01]. In addition, the frequency of C allele was significantly higher in controls compared to SLE patients. Therefore, this allele could have protective effect on SLE susceptibility [OR, 0.7 (95% CI, 0.5 to 0.9); P<0.01] (Table II).
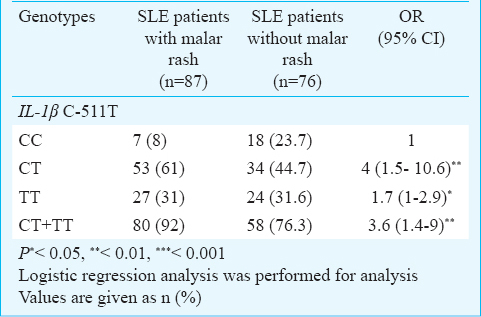
The IL-4 VNTR polymorphism frequency was significantly different between SLE and control groups. Although we did not observe RP2/RP2 genotype in SLE patients and controls, the heterozygous individuals for this polymorphism (RP1/RP2 genotype) were significantly higher in SLE patients and the risk of SLE was 1.8 fold higher in these individuals [OR, 1.8 (95% CI, 1 to 3.1); P<0.05] (Table II). No association was observed between the allelic and genotypic distributions of IL-1β T-31C and IL-4 VNTR polymorphisms and SLE manifestations. However, we observed an association between IL-1β C-511T polymorphism and malar rash and the frequencies of CT and TT genotypes were significantly higher than CC genotype in SLE patients with malar rash compared to SLE patients without this manifestation [OR, 4 (95% CI, 1.5 to 10.6); P<0.01 and OR, 1.7 (95% CI, 1 to 2.9) P<0.05] (Table III).
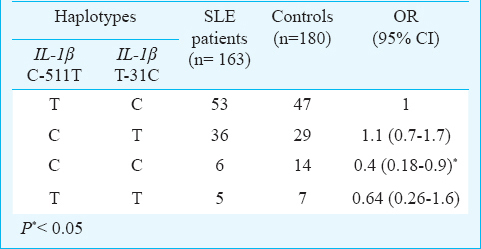
Four haplotypes of the IL-1β consisting of two alleles of each polymorphism are shown in Table IV. The frequency of CC haplotype was significantly lower in SLE patients compared to controls [6 versus 14%, OR, 0.4 (95% CI, 0.18 to 0.9); P=0.015].
Discussion
Our findings showed that CC genotype of IL-1β T-31C polymorphism was significantly lower in SLE patients compared to controls and the risk of SLE was lower in individuals with CC genotype compared to TT genotype. However, no association between IL-1β C-511T polymorphism and SLE was observed. In addition, higher frequencies of RP1/RP2 genotype and RP2 allele of IL-4 VNTR polymorphism were found in SLE group. The frequencies of CT and TT genotypes of IL-1β C-511T polymorphism were significantly higher than CC genotype in SLE patients with malar rash compared to those without this manifestation.
SLE is a complex polygenic disease with immune dysregulation having a critical role in its pathogenesis. In SLE patients like in other autoimmune diseases, cytokines and antibodies are higher than in healthy individuals20. The central role of IL-1β in inflammation suggests that allelic polymorphisms in IL-1β gene might have an effect on susceptibility to SLE. Evidences indicate that T allele of IL-1β T-31C polymorphism is associated with higher production of IL-1β21.
IL-4 as an anti-inflammatory cytokine, mediates the humoral immune response and plays a key role in immune regulation of helper-2 subset of lymphocytes. IL-4 is a key cytokine that induces the activation and differentiation of B cells and is involved in the development of the T cells22. These evidences show that genetic variants that contribute in regulation of IL-1β and IL-4 production may be relevant in genetic susceptibility to SLE.
Muraki et al9 reported that CC and TT genotypes of IL-1β gene C-511T and T-31C polymorphisms were significantly less frequent in SLE patients than controls in Japan, which was in contrast to our findings. However, among African Americans living in the United States, Parks et al23 reported higher risk of SLE in individuals with IL-1β -511T allele than individuals with -511CC genotype, similar to our study this association was not observed in the whites. Tahmasebi et al24 showed no association between C-511T, T-31C polymorphisms and SLE in an Iranian population living in Tehran. However, we found that the TT genotype of T-31C polymorphism was significantly higher in SLE patients in South East of Iran. Wang et al25 performed a meta-analysis (six studies) to determine the effects of IL-1 polymorphisms in susceptibility to SLE and revealed that this polymorphism was not associated with SLE risk in the study subjects. Subgroup analysis for Asian ethnicity indicated that IL-1β C-511T polymorphism was associated with SLE only for TT vs. CT+CC genotypes. In another meta-analysis conducted by Song et al26, no association between IL-1β C-511T polymorphism and SLE was reported.
The information about the association between IL-4 VNTR polymorphism and SLE is scarce. Wu et al13 reported the association between RP1/RP1 genotype and RP1 allele of VNTR and TT genotype and T of C-590T polymorphisms of IL-4 gene with discoid rash in SLE patients in Taiwan. We observed a relation between RP2 allele and RP1/RP2 genotype of IL-4 VNTR polymorphism and SLE susceptibility.
It was suggested that these differences in the genotypes and alleles distribution might be due to different sample size and different selection criteria adopted for patients and controls in particular clinical manifestations, ethnicity and environmental risk factors. As mentioned previously by Manchanda et al27, there is considerable evidence that polymorphisms in the regulatory regions of cytokine genes are affected by ethnicity, therefore, these differences could be related to ethnic and clinical heterogeneity between populations.
Our study had certain limitations. The first was the low sample size, and second that environmental conditions which are known to have essential role in SLE initiation and progression, were not studied. Therefore, further investigations using a larger sample size with effect of environmental conditions and different ethnic groups are necessary to confirm the present results.
In conclusion, our study showed an association between IL-1β T-31C and IL-4 VNTR polymorphisms and SLE susceptibility. There was no association between IL-1β C-511T polymorphism and SLE. Further, T allele of IL-1β C-511T polymorphism could be related with malar rash manifestation in SLE patients.
Acknowledgment
The authors thank Zahedan Deputy of Research Affairs Zahedan University of Medical Sciences, Iran, for funding this project.
Conflicts of Interest: None.
References
- Association of IRF5 in UK SLE families identifies a variant involved in polyadenylation. Hum Mol Gen. 2007;16:579-91.
- [Google Scholar]
- Deoxyribonuclease I gene polymorphism and susceptibility to systemic lupus erythematosus. Clin Rheumatol. 2016;35:101-5.
- [Google Scholar]
- The United States to Africa lupus prevalence gradient revisited. Lupus. 2011;20:1095-103.
- [Google Scholar]
- Prevalence of musculoskeletal disorders in southeastern Iran: a WHO-ILAR COPCORD study (stage 1, urban study) Int J Rheum Dis. 2013;16:509-17.
- [Google Scholar]
- Genetic factors predisposing to systemic lupus erythematosus and lupus nephritis. Semin Nephrol. 2010;30:164-76.
- [Google Scholar]
- Delineating the genetic basis of systemic lupus erythematosus. Immunity. 2001;15:397-408.
- [Google Scholar]
- Systemic autoimmune disease arises from polyclonal B cell activation. J Exp Med. 1987;165:1755-60.
- [Google Scholar]
- Elevation of proinflammatory cytokine (IL-18, IL-17, IL-12) and Th2 cytokine (IL-4) concentrations in patients with systemic lupus erythematosus. Lupus. 2000;9:589-93.
- [Google Scholar]
- Polymorphisms of IL-1 beta gene in Japanese patients with Sjögren's syndrome and systemic lupus erythematosus. J Rheumatol. 2004;31:720-5.
- [Google Scholar]
- Four new members expand the interleukin-1 superfamily. J Biol Chem. 2000;275:1169-75.
- [Google Scholar]
- IL-4 haplotype -590T, -34T and intron-3 VNTR R2 is associated with reduced malaria risk among ancestral indian tribal populations. PloS One. 2012;7:e48136.
- [Google Scholar]
- The human interleukin-1 receptor antagonist (IL1RN) gene is located in the chromosome 2q14 region. Genomics. 1993;15:173-6.
- [Google Scholar]
- Polymorphisms of the interleukin-4 gene in Chinese patients with systemic lupus erythematosus in Taiwan. Lupus. 2003;12:21-5.
- [Google Scholar]
- Updating the American College of Rheumatology revised criteria for the classification of systemic lupus erythematosus. Arthritis Rheum. 1997;40:1725.
- [Google Scholar]
- Modified salting-out method: high-yield, high-quality genomic DNA extraction from whole blood using laundry detergent. J Clin Lab Anal. 2005;19:229-32.
- [Google Scholar]
- Association analysis between variants of the interleukin-1beta and the interleukin-1 receptor antagonist gene and antidepressant treatment response in major depression. Neuropsychiatr Dis Treat. 2008;4:269-76.
- [Google Scholar]
- Association of interleukin 1B gene polymorphism and gastric cancers in high and low prevalence regions in China. Gut. 2003;52:1684-9.
- [Google Scholar]
- Interleukin-1beta, interleukin-1 receptor antagonist, interleukin-4, and interleukin-10 gene polymorphisms: relationship to occurrence and severity of rheumatoid arthritis. Arthritis Rheum. 1999;42:1093-100.
- [Google Scholar]
- Cubic exact solutions for the estimation of pairwise haplotype frequencies: implications for linkage disequilibrium analyses and a web tool ‘CubeX’. BMC Bioinformatics. 2007;8:428.
- [Google Scholar]
- Serum Levels of IL-18 in Iranian females with systemic lupus erythematosus. Medicinski Arhiv. 2013;67:237-40.
- [Google Scholar]
- Interleukin-1, interleukin-1 receptors and interleukin-1 receptor antagonist. Int Rev Immunol. 1998;16:457-99.
- [Google Scholar]
- Possible association of IL-4 VNTR polymorphism with susceptibility to preeclampsia. Biomed Res Int 2014 2014:497031.
- [Google Scholar]
- Systemic lupus erythematosus and genetic variation in the interleukin 1 gene cluster: a population based study in the southeastern United States. Ann Rheum Dis. 2004;63:91-4.
- [Google Scholar]
- Interleukin-1 gene cluster and IL-1 receptor polymorphisms in Iranian patients with systemic lupus erythematosus. Rheumatol Int. 2013;33:2591-6.
- [Google Scholar]
- The association of IL1alpha and IL1beta polymorphisms with susceptibility to systemic lupus erythematosus: a meta-analysis. Gene. 2013;527:95-101.
- [Google Scholar]
- Associations between interleukin 1 polymorphisms and susceptibility to systemic lupus erythematosus: a meta-analysis. Hum Immunol. 2014;75:105-12.
- [Google Scholar]
- Ethnicity greatly influences the interleukin-1 gene cluster(IL-1b promoter, exon-5 and IL-1Ra) polymorphisms: a pilot study of a north Indian population. Asian Pac J Cancer Prev. 2005;6:541-6.
- [Google Scholar]






