Translate this page into:
In-house, simple & economical phage technique for rapid detection of rifampicin, isoniazid, ethambutol, streptomycin & ciprofloxacin drug resistance using Mycobacterium tuberculosis isolates
Reprint requests: Dr D.S. Chitnis, Department of Microbiology, Immunology & Molecular biology, Intermediate Referral Laboratory for Mycobacteriology, Choithram Hospital & Research Centre, Manik Bagh Road, Indore 452 014, India e-mail: ds_chitnis@rediffmail.com
-
Accepted: ,
This is an open-access article distributed under the terms of the Creative Commons Attribution-Noncommercial-Share Alike 3.0 Unported, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
This article was originally published by Medknow Publications & Media Pvt Ltd and was migrated to Scientific Scholar after the change of Publisher.
Abstract
Background & objectives:
Multiple drug resistance (MDR) among Mycobacterium tuberculosis poses a serious therapeutic problem. Early detection of MDR can be valuable but the conventional drug susceptibility tests take 4-6 wk time after the laboratory isolation of M. tuberculosis. The bacterial phage assay has been reported as a rapid tool for rifampicin susceptibility testing of tubercle bacilli using the suspension of isolated cultures. The present study was aimed to set up a phage assay for testing drug susceptibility to isoniazid (INH), rifampicin, ethambutol, streptomycin and ciprofloxacin in M. tuberculosis isolates.
Methods:
Mueller-Hinton broth instead of Middle Brook 7H9 broth was used to make it more economical. The phage assay was compared with the proportion method using 100 M. tuberculosis isolates from pulmonery TB cases. Phage assay results were available in 48 h for rifampicin and streptomycin while 72 h required for INH, ethambutol and ciprofloxacin. The assay was compared with gold standard proportion method. Interpretation of the results was easy and clear.
Results:
In the present study, sensitivity and specificity of the phage assay when compared to proportion method were in the range of 97 to 100 per cent for all the drugs except for ciprofloxacin for which it was 93 and 96 per cent, respectively.
Interpretation & conclusions:
The phage assay was economic, easy to perform and rapid for the detection of drug resistance in M. tuberculosis isolates with no requirement of expensive equipment. It is within the reach of microbiology laboratories in developing countries having high loads of tuberculosis.
Keywords
Drug resistance
mycobacteriophage
MDR
sensitivity
specificity
tuberculosis
The emergence of multidrug-resistant tuberculosis (MDR-TB) and, more recently, extensively drug-resistant TB (XDR-TB) is a major threat to global TB control programmes12.
Multi drug-resistant tuberculosis (MDR-TB), caused by isolates resistant to at least isoniazid (INH) and rifampicin, forms an alarming problem for the successful management of tuberculosis3. The prevalence rate of MDR-TB in India is around 3 per cent among new cases, 12 per cent among pretreatment cases4 and an alarming 17 per cent among patients who have been treated earlier and developed multi-drug resistance (MDR). MDR-TB4 has been reported from almost all parts of the world, primarily as a consequence of poor and irregular treatment services, resulting into increased costs towards treatment and also increased risk of transmission of these resistant strains of the bacilli.
Rapid detection of MDR-TB can help in the effective management of such cases. The DOT plus programme is being geared up in India and other countries with high loads of TB5. However, the current phenotypic methods of assessing drug susceptibility for Mycobacterium tuberculosis are slow and take 4-6 wk after laboratory isolation of the organisms. Rapid methods based on polymerase chain reaction (PCR) are considered as alternative approach for the detection of MDR but detection of only rifampicin resistance through rpoB gene mutation6–9 has gained confidence, while in case of INH variations in resistance gene pose a problem. Mycobacteriophage-based techniques have also been reported for detection of viable bacilli in clinical specimens1011 as well as for antimicrobial susceptibility testing12–22.
The mycobacteriophage-based assays depend upon the ability of resistant mycobacteria to support phage replication after being exposed to drugs, while sensitive bacilli get inactivated and hence are not able to support phage infection. Extracellular phages are inactivated with a virucidal agent, whereas intracellular phages are protected and replicate, causing their lysis and the release of a new phage progeny detected by the production of plaques on a fast-growing M. smegmatis (indicator strain) lawn. The released phages can be visualized on indicator plates as clear areas (plaques) within a lawn of fast growing M. smegmatis12. This ingenious method does not require specialized laboratory skills or sophisticated equipment.
The aim of the present study was to set up a phage assay for drug susceptibility to isoniazid, rifampicin, ethambutol, streptomycin and ciprofloxacin using suspension of M. tuberculosis isolates and to compare it with proportion method.
Material & Methods
Mycobacterium isolates: A total of 100 consecutive M. tuberculosis isolates from respiratory samples of pulmonary tuberculosis cases referred to Microbiology Department of Choithram Hospital and Research Centre (CHRC), Indore, India, during January 2009 to September 2010 were included in the study. The study protocol was approved by the Research and Ethical Committee of CHRC, Indore. The standard strain of M. tuberculosis H37Rv, and M. tuberculosis wild isolates resistant to the drugs and maintained in the laboratory were used for quality control of every batch. M. tuberculosis was identified by using standard biochemical tests23 and 3-4 wk old culture on Lowenstein-Jensen Medium (LJ) was used for making suspensions for the drug susceptibility tests.
Standardization of phage assay: Extensive standardization work was carried out to optimize the following conditions: liquid medium for cultivation of M. smegmatis; evaluation of various solid media for phage assay; inoculum size M. smegmatis for indicator plate; density of M. tuberculosis suspension; cut-off for plaque counts for sensitive and resistant M. tuberculosis; optimal concentration of drugs for exposure to M. tuberculosis; drug exposure time for M. tuberculosis; and concentration and exposure time period of ferrous ammonium sulphate (FAS) for killing extracellular phages.
Adequate controls replicated were included for the standardization work to fix up the optimal conditions.
Phage assay protocol for drug susceptibility testing: The suspension of M. tuberculosis isolates was prepared in Muller-Hinton (M-H) broth in plastic tubes containing 8-10 glass beads. The inoculum was adjusted to McFarland tube No.1. INH, ethambutol and streptomycin (Sigma-Aldrich Chemicals GmbH, Steinheim, Germany) stock solutions (10 mg/10 ml) were made in distilled water and filtered through 0.2 μ pore size membrane. Rifampicin and ciprofloxacin stock solutions were made in dimethyl formamide (Sigma, USA). All stock solutions were aliquoted and preserved at -70°C till use.
Working solution was prepared in M-H broth (B.D. Difco, USA) The drug concentrations used were: INH (0.4 μg/ml), rifampicin (2 μg/ml), ethambutol (16 μg/ml), streptomycin (2 μg/ml), and ciprofloxacin (4 μg/ml). Plain M-H broth without drug served as control.
M-H agar supplemented with 0.4 per cent glycerol and 1 per cent glucose was used for plate pouring in phage assay. M. smegmatis mc2155 strain was used for propagation of mycobacteriophages on M-H agar. Mycobacteriophage (D29) was propagated on M. smegmatis, as described earlier24. The supernatant containing phages was passed through 0.22 μ Millipore filter membrane and stored at -20°C. The phage titre was determined using a standard spot test24.
M. tuberculosis isolates were exposed to rifampicin for 24 h while exposure time for other drugs was 48 h (based on our preliminary work and other studies)11–21. The concentrations of the drugs used for phage assay were also based on our preliminary study (unpublished). The assay was carried out in 10 ml plastic screw capped tubes (Nunc, USA). Five hundred microlitre of M. tuberculosis suspension was mixed with 500 μl drug. Wild drug resistant strains (laboratory isolates) and M. tuberculosis H37Rv were included in all batches. The tubes were incubated for 24/48 h at 37°C.
Mycobacteriophage D29 (200 μl, 108 pfu /ml) was added into the respective tubes and incubated for 90 min at 37°C. The extracellular phages were inactivated with 200 μl of phagicidal agent ferrous ammonium sulphate (working concentration 30mM) (Merck, India) with contact time of 10 min. The contents of the tubes were transferred to 10 ml M-H broth in18 × 150 mm tube and vortexed. One ml of the broth mixture and 1 ml of M. smegmatis suspension were dispensed in sterile disposable Petri dish (90 mm); and 10 ml of molten M-H agar supplemented with glycerol and glucose was added and mixed by rotating clock-wise and anti clock-wise. The plates were left for 10 min for solidification and transferred to incubator at 37°C for over night. Results were analyzed by counting the plaques. In control plates the plaque count ranged from 90-150 on plate. The isolates were considered resistant if reduction in plaque count was less than 80 per cent compared to control plate, and susceptible if more than 80 per cent reduction was observed.
Drug susceptibility testing by proportion method: The proportion method25 was carried out using L-J medium. The recommended critical concentrations25 of INH 0.2 μg/ml, rifampicin 40 μg/ml, ethambutol 2 μg/ml, streptomycin 4 μg/ml and ciprofloxacin 2 μg/ml were used. The first reading was taken after 28 days of incubation and the second on 42nd day. The percentage resistance (R) was calculated as the ratio of the number of colonies on the drug containing media to those on the control medium.
Statistical analysis: Data were analysed using Bayes-Theorem, 2×2 contingency tables to calculate sensitivity, specificity, positive predictive value (PPV) and negative predictive value (NPV) using StatsDirect statistical software version 2.7.2, UK. McNemar's test was applied using Graphpad software Inc., USA (two tailed pair test) where each isolate was matched for phage assay and proportion method.
Results
Of the 100 M. tuberculosis isolates tested, 58 (58%) were resistant to INH, 43 to rifampicin, 55 isolates streptomycin, 32 to ethambutol and 28 isolates were resistant to ciprofloxacin by proportion method. Forty isolates were resistant to both rifampicin and INH (i.e. MDR) by proportion method.
When phage assay was compared with proportion method, 57 of 58 INH resistant isolates and 41 of the 42 sensitive isolates were correctly identified by the phage assay (Table). Discordance for phage assay was seen among only two isolates. The sensitivity of the mycobacteriophage assay in detecting INH resistance was 98 per cent; and the specificity achieved was also 98 per cent. Both positive and negative predictive values were 98 per cent.
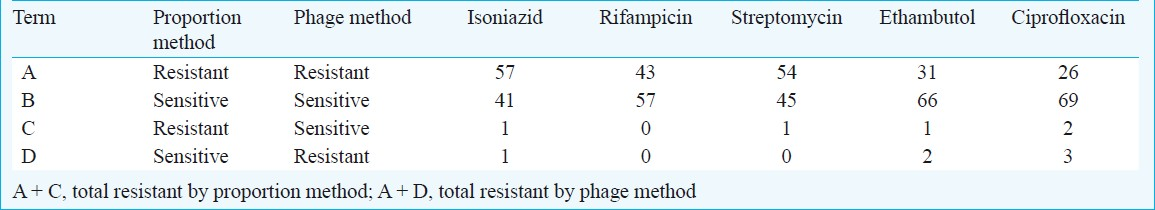
In case of rifampicin, both resistant and sensitive isolates were correctly identified by the phage assay. The sensitivity of the mycobacteriophage assay in detecting rifampicin resistance was 100 per cent; and the specificity achieved was also 100 per cent. Positive and negative predictive values were also 100 per cent for both. The sensitivity and specificity for ethambutol were 97 per cent for both. Positive and negative predictive values were 94 and 99 per cent, respectively. The sensitivity and specificity for ciprofloxacin were 93 and 96 per cent, respectively. The positive and negative predictive values were 90 and 97 per cent, respectively. Each isolate was checked for drug sensitivity by both conventional proportion method and phage assay and thus, matched pairs were formed to allow McNemar's test.
Discussion
The phage based assay was developed by Wilson and colleagues22, and was applied to rifampicin and isoniazid susceptibility testing for clinical isolates of M. tuberculosis. The commercial phage assay is available only for detection of resistance for rifampicin2627. Hence, the present study was aimed to standardize in-house phage assay for all the first line anti-TB drug resistance detection. Fluoroquinolones are widely used along with other anti-TB drugs and therefore, were also included in the present study.
Middle brook 7H9 medium was replaced with Mueller Hinton broth to make the assay economical. The cost of the commercial phage assay (FASTPlaque TB-RIF, Biotec Laboratories Ltd, UK) for rifampicin resistance is over 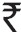 500 per test while the cost of consumables for in-house phage based assay was less than
500 per test while the cost of consumables for in-house phage based assay was less than 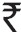 50 per drug per isolate.
50 per drug per isolate.
In the present study, sensitivity and specificity of the phage assay when compared to proportion method were in the range of 97 to 100 per cent and for ciprofloxacin it was 93 and 96 per cent, respectively. The data on positive and negative predictive values, favoured the utility of the in-house phage assay.
Reduction in plaque count >80 per cent was considered as susceptible to the drug in the present study. However, in case of rifampicin and streptomycin susceptible isolates, more than 99 per cent reduction in plaque count was observed. This appeared to be due to high and rapid bactericidal activity of the drugs for Mycobacterium. Another mycobacteriophage-based assay, the luciferase reporter phage (LRP) assay showed a 99 per cent reduction in the light signal after an overnight incubation of susceptible isolates with streptomycin28. These two drugs required shorter time exposure (24 h) compared to other drugs which required 48 h exposure as observed in the other study19. The mode of action of the drug may also be a contributory factor, e.g. activity at transcription/translation level (streptomycin, rifampicin) and at cell wall level (INH, ethambutol)29.
Our findings were comparable with other studies for rifampicin1415172030, INH and or streptomycin12161821. In case of fluoroquinolones, only two studies reported phage assay1231, and information is scanty for ethambutol11.
Molecular methods have limitations since the PCR is directed towards the particular mutant. Such difficulty does not arise for phage based assays since these are dependent on the viability following drug exposure.
The drug resistance determination on direct clinical samples such as sputum will be of immense value, since it will bring down the reporting time to 2-3 days on the direct samples.
In conclusion, the in-house phage assay standardized for detection of drug resistance in M. tuberculosis culture suspension showed high sensitivity and specificity and the results were available within 2-3 days. The assay was economical and within the reach of microbiology laboratories in developing countries. Overall concordance with proportion method was good.
Acknowledgment
This work was supported by young scientist scheme grant SR/FT/L-116/2006 from DST (Department of Science and Technology), New Delhi, India.
References
- Multidrug-resistant and extensively drug-resistant Mycobacterium tuberculosis: epidemiology and control. Expert Rev Anti Infect Ther. 2007;5:857-71.
- [Google Scholar]
- Worldwide emergence of extensively drug-resistant tuberculosis. Emerg Infect Dis. 2007;13:380-7.
- [Google Scholar]
- World Health Organization. In: Global tuberculosis control: epidemiology, strategy, financing: WHO Report 2009. Geneva: World Health Organization; 2009. (WHO /HTM/TB/2009.411)
- [Google Scholar]
- A prioritised research agenda for DOTS-Plus for multidrug-resistant tuberculosis (MDRTB) Int J Tuberc Lung Dis. 2003;7:410-4.
- [Google Scholar]
- Contribution of rpoB mutations to development of rifamycin cross-resistance in Mycobacterium tuberculosis. Antimicrob Agents Chemother. 1998;42:1853-7.
- [Google Scholar]
- Evaluation of a polymerase chain reaction-based universal heteroduplex generator assay for direct detection of rifampin susceptibility of Mycobacterium tuberculosis from sputum specimens. Clin Infect Dis. 1998;26:446-50.
- [Google Scholar]
- Detection of rifampicin-resistance mutations in Mycobacterium tuberculosis. Lancet. 1993;341:647-50.
- [Google Scholar]
- Mutations in the rpoB gene of rifampin-resistant Mycobacterium tuberculosis isolates in Spain and their rapid detection by PCR-enzyme-linked immunosorbent assay. J Clin Microbiol. 2001;39:1813-8.
- [Google Scholar]
- Use of a phage-based assay for phenotypic detection of mycobacteria directly from sputum. J Clin Microbiol. 2003;41:680-8.
- [Google Scholar]
- Study of phage based diagnostic technique for tuberculosis. Indian J Tuberc. 2007;54:36-40.
- [Google Scholar]
- Evaluation of a bacteriophage-based assay (phage amplified biologically assay) as a rapid screen for resistance to isoniazid, ethambutol, streptomycin, pyrazinamide, and ciprofloxacin among clinical isolates of Mycobacterium tuberculosis. J Clin Microbiol. 1999;37:3528-32.
- [Google Scholar]
- Use of a mycobacteriophage-based assay for rapid assessment of susceptibilities of Mycobacterium tuberculosis isolates to isoniazid and influence of resistance level on assay performance. J Clin Microbiol. 2006;44:201-5.
- [Google Scholar]
- Utility of an in-house mycobacteriophage-based assay for rapid detection of rifampin resistance in Mycobacterium tuberculosis clinical isolates. J Clin Microbiol. 2003;41:2647-9.
- [Google Scholar]
- Evaluation of phage assay for rapid phenotypic detection of rifampicin resistance in Mycobacterium tuberculosis. Ann Clin Microbiol Antimicrob. 2006;5:5-11.
- [Google Scholar]
- Evaluation of rifampicin and isoniazid susceptibility testing of Mycobacterium tuberculosis by a mycobacteriophage D29-based assay. J Med Microbiol. 2007;56:360-4.
- [Google Scholar]
- Low-cost rapid detection of rifampicin resistant tuberculosis using bacteriophage in Kampala, Uganda. Ann Clin Microbiol Antimicrob. 2007;6:1.
- [Google Scholar]
- Comparison of redox and D29 phage methods for detection of isoniazid and rifampicin resistance in Mycobacterium tuberculosis. Clin Microbiol Infect. 2006;12:293-6.
- [Google Scholar]
- Evaluation of reverse transcription-PCR and a bacteriophage-based assay for rapid phenotypic detection of rifampin resistance in clinical isolates of Mycobacterium tuberculosis. J Clin Microbiol. 1999;37:3524-7.
- [Google Scholar]
- Colorimetric phage-based assay for detection of rifampin-resistant Mycobacterium tuberculosis. J Clin Microbiol. 2007;45:1330-2.
- [Google Scholar]
- Rapid screening of Mycobacterium tuberculosis for susceptibility to rifampicin and streptomycin. Int J Tuberc Lung Dis. 2000;4:69-75.
- [Google Scholar]
- Evaluation of a new rapid bacteriophage-based method for the drug susceptibility testing of Mycobacterium tuberculosis. Nat Med. 1997;3:465-8.
- [Google Scholar]
- Koneman EW, Allen SD, Janda WM, Schreckenberger PC, Winn EC Jr, eds. Color atlas and textbook of diagnostic microbiology (5th ed). Philadelphia: JB Lippincott; 1997.
- Mycobacteria: laboratory methods for testing drug sensitivity and resistance. Bull World Health Organ. 1963;29:565-78.
- [Google Scholar]
- Evaluation of FASTPlaque TB-RIF, a rapid, manual test for the determination of rifampicin resistance from Mycobacterium tuberculosis cultures. Int J Tuberc Lung Dis. 2001;5:906-11.
- [Google Scholar]
- Simple, phage-based (FASTPplaque) technology to determine rifampicin resistance of Mycobacterium tuberculosis directly from sputum. Int J Tuberc Lung Dis. 2004;8:1114-9.
- [Google Scholar]
- The use of luciferase-reporter phage for antibiotic-susceptibility testing of mycobacteria. Methods Mol Biol. 1998;101:431-55.
- [Google Scholar]
- Rapid assessment of drug susceptibilities of Mycobacterium tuberculosis by means of luciferase reporter phages. Science. 1993;260:819-22.
- [Google Scholar]
- In-house phage amplification assay is a sound alternative for detecting rifampin-resistant Mycobacterium tuberculosis in low-resource settings. Antimicrob Agents Chemother. 2005;49:425-7.
- [Google Scholar]
- Comparison of the performance of two mycobacteriophage D29-based protocols for fluoroquinolone susceptibility testing in Mycobacterium tuberculosis. J Microbiol Methods. 2009;79:371-3.
- [Google Scholar]






