Translate this page into:
Identifying potential pitfalls in interpreting mitochondrial DNA mutations of male infertility cases
Reprint requests: Dr Malliya Gounder Palanichamy, Laboratory for Conservation & Utilization of Bio-resources, Yunnan University, Kunming 650 091, Yunnan, PR China e-mail: empalani@yahoo.com
-
Received: ,
This is an open-access article distributed under the terms of the Creative Commons Attribution-Noncommercial-Share Alike 3.0 Unported, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
This article was originally published by Medknow Publications & Media Pvt Ltd and was migrated to Scientific Scholar after the change of Publisher.
Abstract
Background & objectives:
Recently, a significantly higher ratio of nucleotide changes in the mtDNA genes: COII, ATPase 6, ATPase 8, ND2, ND3, ND4, and ND5 was reported in spermatozoa from populations of infertile Indian men, compared suggesting that screening for mtDNA mutations could provide insight into the aetiology of male infertility. In this study, we examined the published data and found serious errors in the original acquisition and analysis of the data.
Methods:
The mtDNA data associated with male infertility in Indian populations were retrieved from the published sources. The mtDNA substitution values of infertile and control groups were evaluated using phylogenetic methods and previously published mtDNA phylogenies.
Results:
Most of the mtDNA polymorphisms reported as significantly correlated with infertility were more commonly found in general populations. Further, our analysis showed that some of the mtDNA substitutions were erroneously overestimated in the infertile groups and underestimated in the control groups, and vice-versa.
Interpretation & conclusions:
Contrary to earlier claims, our analysis demonstrated no significant association between the mtDNA polymorphisms and male infertility in these studies. Further, these errors in the published data impune the usefulness of mitochondrial molecular analyses in male infertility diagnosis.
Keywords
Male infertility
mitochondrial DNA mutation
spermatozoa
The traditional semen parameters in the diagnosis of male infertility are primarily sperm concentration, motility and morphology, and semen volume. In the past few years however, researchers have turned to genetic analysis and a possible link between mitochondrial DNA (mtDNA) mutations and male infertility has been suggested12. Multiple studies have shown that large numbers of infertile men possess mutations in mitochondrial DNA in genes regulating oxidative phosphorylation system2. It has been speculated that high incidence of these kinds of mtDNA mutations in sperm lead to decreased energy generation and result in severely impaired sperm motility and subsequently lead to diminished fertility3. However, the actual incidence of mtDNA mutations associated with sperm motility is still a matter of debate; some groups find the link between sperm motility and mtDNA variants14–9, while others have not found such an association10–14.
Thangaraj et al3 have analyzed the sequence variation of mtDNA in spermatozoa of an oligoasthenotetratozoospermia patient and found as many as nine missense and 27 silent mutations as well as a 2-bp deletion, clustering in the region 6241-9167. Recently, Kumar and colleagues7–91516 have observed a high frequency of nucleotide changes in the mitochondrial genes COII, ATPase 6, ATPase 8, ND2, ND3, ND4, and ND5 in the semen/blood cells of infertile men. These studies suggest that mtDNA plays a crucial role in sperm dysfunction and further suggest it serve as a potential diagnostic marker in infertile men, especially in cases of idiopathic oligoasthenozoospermia.
In contrast, Bandelt13 reported that previous screening of mtDNA data from male infertility cohort studies contain obvious errors and give false association with infertility. Pereira et al11 presented strong evidence against the accepted claim of a major role played by mtDNA in male infertility. The possible role of the mitochondria in spermatogenesis, which may contribute to male infertility, has also been discussed17.
In view of certain flaws in the previously published data, we reanalyzed some current published mtDNA data associated with male infertility and evaluated the pathogenic status of mtDNA mutation.
Material & Methods
The mtDNA data associated with male infertility in Indian population published by Thangaraj3, and Kumar & colleagues7–91516 were reanalysed. The high incidence mtDNA single nucleotide substitutions variables related to infertility cases and controls were assessed using the mtDNA phylogeny, which guides to assist the evolutionary pathways and may help to identify the discordant features of the recorded mutations. Since the mtDNA sequences in the studies considered here are from Indian ancestry, we referred to the south Asian mtDNA phylogenies18–20.
Results & Discussion
Abnormal mtDNA sequence polymorphism in an oligoasthenotetratozoospermia case: An unusual profile and high prevalence of mtDNA mutations in sperms have been asserted by Thangaraj et al3 in a single case of oligoasthenoteratozoospermia (OAT). The authors targeted their analysis on COI, COII, ATPase6, ATPase8, tRNA serine I, tRNA lysine, and ND3 of the sperm's mtDNA of a man with OAT and they found as many as nine missense and 27 silent mutations as well as a 2-nucleotide deletion in the COII gene.
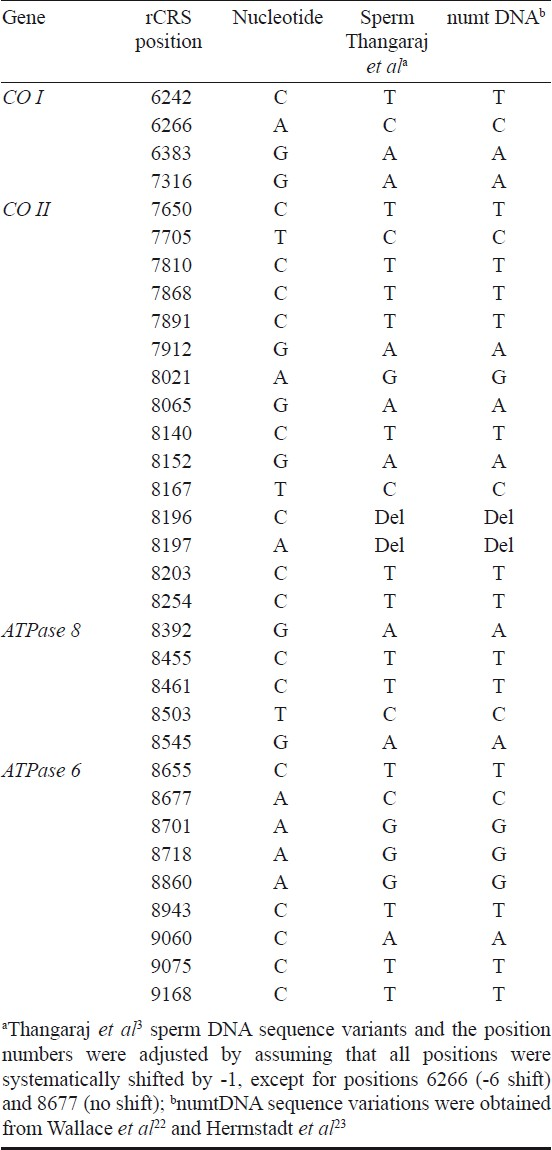
When we examined their data carefully we found that the listed nucleotides in the reference sequence do not correspond to the nucleotides of the revised Cambridge Reference Sequence (rCRS)21; instead, by mistake, there was a base shifted from the rCRS by -1 nucleotide position. Once their sequence was properly aligned and the nucleotide variants were correctly assigned, a comparison with the published data showed that the authors erroneously amplified and sequenced the nuclear mitochondrial DNA-like sequence (numtDNA) located on chromosome I.
Of the 36 nucleotide substitutions listed related to OAT case, 31 sites coincide with the numtDNA sequence (Table I). In addition, the two deleted bases in COII gene of the OAT case were also found in numtDNA sequence. With the vast majority of the patient's variants coinciding with the numt sequence it is safe to say that this patient had only five substitutions, and those in the numt sequence, not the mtDNA. Here a minor misalignment resulted in a major error in the interpretation of the data; yet, this paper has been quoted many times by many other authorities in the field.
Misdocumented mtDNA mutations in oligoasthenozoospermic cases: Kumar et al7 have reported a high incidence of some single nucleotide substitutions (SNPs) in the mtDNA genome of men with asthenospermia. They screened the mtDNA mutations in 23 oligoasthenozoospermia (OA) cases and found a total of 22 substitutions. Of the 22 nucleotide changes, two nucleotides- 8701 and 8860 were found in 12 and 20 patients, respectively. In another study15, the same research group analyzed the semen and blood samples of 25 OA cases and 20 controls and found the polymorphisms- 750, 4769, and 8860 in all cases but these mutations were detected only in 12 controls. In addition, another nucleotide substitution 11719 was detected in sperm DNA samples of 19 cases whereas only 14 samples of blood DNA had this mutation15. The mtDNA mutations analysis was repeated by the authors using another group of individuals (33 OA cases and 25 controls) and they found similar results16. Altogether, these authors concluded that the mtDNA mutations- 750, 4769, 8701, 8860, and 11719 were more frequent in men with OA than in controls1516. When their data were compared with mtDNA data from a large Indian population, it was obvious that the polymorphisms they found were widespread in the general populace18–20. This result indicates that either OA is common and widespread in the Indian populace as a whole or the authors have, by dint of a small sample size and the oversight of not examining the available data on polymorphisms, discovered an incorrect correlation and come to false conclusions.
Recently, the same group showed significant nucleotide changes in the mitochondrial gene: ATPase6, ATPase8, ND2, ND3, ND4, and ND5 in the semen of the OA groups compared to controls9. The authors suggest that screening the mtDNA mutations can give some insight into the aetiology of motility disorders in OA men. However, closer examination of their Table III data revealed many errors and thus cannot support the hypothesis that mutant mtDNA plays a role in the impaired fertility of OA men. For example, transition of nucleotide A to G at site 4769 is shared by virtually all mtDNAs worldwide that are not closest relatives of the rCRS21 (rCRS sequence belong to H haplogroup). In their Table III9, the frequency was recorded as 4/33 and 23/30 in infertile and control groups, respectively. If one assumes this mutation frequency in the infertile and control groups, then one would conclude that remaining 88 per cent (29/33) members of infertile and 23 per cent (7/30) of control group belong to haplogroup H. We never observed such a group in India. Moreover, it contradicts their previous report, that the mutation 4769 is observed more frequently in men with OA than in controls16. In addition, the most common nucleotide substitution is A8860G, this mutation is generally shared by all individuals outside haplogroup H2a2a, while most authors erroneously detected in less than half (14/30) of the control group. The nucleotide-haplogroup analysis results are presented in Table II.

The mutation G5400C highly occurred (70%) in control group9, which is not found in the Indian published database (1059 complete mtDNA genome sequences)18–20 as well as our unpublished (300 complete mtDNA sequences) sequences, so we suspect existence of this mutation in the control population. The remaining four variants, C8394T, C10165T, C10207T, and G13708A were associated significantly with infertile group. The mutation G13708A a notably major mutation hot spot site in the mtDNA has been found in different India specific M, N, and R derived haplogroups background. It is, therefore, doubtful that this mutation is actually associated with infertility. Thus, the three remaining variants may be interesting; nevertheless, further investigation will be necessary to substantiate or refute the claims.
The substitution G11719A was generally found in all mtDNA sequence except West Eurasian haplogroup R0. According to their frequency in control group, half of the mtDNAs did not have this polymorphism, it means that 50 per cent of control mtDNAs belonged to members of haplogroup R0 (Table II). Such a high frequency R0 population samples has never been observed in India. The mutations A8701G and G10398A correspond to two major Asian haplogroup M and N. These variants were reported 48 per cent (M) and 76 per cent (N) in infertile and 87 per cent (M) and 80 per cent (N) in control groups. The fact that all Indian mtDNA lineages were derived from macrohaplogroup M and N, under this condition one should observe the total frequencies equivalent to 100 per cent (frequency of M + frequency of N = 100%), but the reported frequencies in infertile and control groups exceeded this total value.
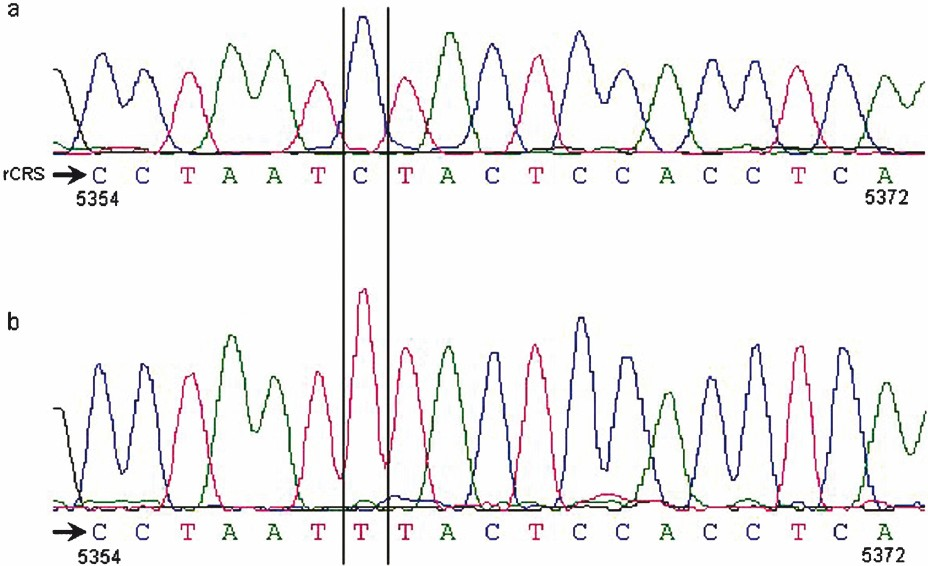
Flawed electropherogram: Shamsi et al8 reported a transition T12705C in single OAT case. In addition, related to this transition they also provided an electropherogram image. By inspecting their diagram two obvious flaws were found: the first was that the pointed electropherogram peak was corresponding to nucleotide base T but the authors erroneously typed in to base C. The second flaw was that the electropherogram representing 19 nucleotides was actually corresponding to nucleotide positions 5354 to 5372. In fact, the marked nucleotide differed from the rCRS at 5360 site, which is one of the defining markers for U7 haplogroup.
In conclusion, none of the data reanalyzed in the present study demonstrate a link between male infertility in Indian populations and mtDNA changes. Identifying mtDNA polymorphisms related to male infertility could be very helpful, but data should be collected, analyzed, and interpreted with special care. It is emphasized that careful assessment of identified mtDNA polymorphisms must be undertaken in clinical male infertility cases.
References
- The impact of mitochondrial genetics on male infertility. Int J Androl. 2005;28:65-73.
- [Google Scholar]
- Role of reactive oxygen species in the pathogenesis of mitochondrial DNA (mtDNA) mutations in male infertility. Indian J Med Res. 2009;129:127-37.
- [Google Scholar]
- Sperm mitochondrial mutations as a cause of low sperm motility. J Androl. 2003;24:388-92.
- [Google Scholar]
- High incidence of single nucleotide substitutions in the mitochondrial genome is associated with poor semen parameters in men. Int J Androl. 2001;24:175-82.
- [Google Scholar]
- Can mitochondrial DNA mutations cause sperm dysfunction? Mol Hum Reprod. 2002;8:719-21.
- [Google Scholar]
- Necessity of nuclear and mitochondrial genome analysis prior to assisted reproductive techniques/intracytoplasmic sperm injection. Indian J Biochem Biophys. 2007;44:437-42.
- [Google Scholar]
- Mitochondrial DNA mutations in etiopathogenesis of male infertility. Indian J Urol. 2008;24:150-4.
- [Google Scholar]
- Oxidative stress and sperm mitochondrial DNA mutation in idiopathic oligoasthenozoospermic men. Indian J Biochem Biophys. 2009;46:172-7.
- [Google Scholar]
- Human mtDNA haplogroups and reduced male fertility: real association or hidden population substructuring. Int J Androl. 2005;28:241-7.
- [Google Scholar]
- No evidence for an mtDNA role in sperm motility: data from complete sequencing of asthenozoospermic males. Mol Biol Evol. 2007;24:868-74.
- [Google Scholar]
- Mutation C11994T in the mitochondrial ND4 gene is not a cause of low sperm motility in Portugal. Fertil Steril. 2008;89:738-41.
- [Google Scholar]
- Misanalysis gave false association of mtDNA mutations with infertility. Int J Androl. 2008;31:450-3.
- [Google Scholar]
- Correlation of the 4977 bp mitochondrial DNA deletion with human sperm dysfunction. BMC Res Notes. 2009;2:18.
- [Google Scholar]
- Mitochondrial DNA mutations and polymorphism in idiopathic asthenozoospermic men of Indian origin. In: Proceedings of the 57th Annual Meeting of American Society of Human Genetics. 2007.
- [Google Scholar]
- Increased lipid peroxidation, low antioxidant levels and nucleotide changes in mitochondrial DNA in idiopathic asthenozoospermic men of Indian origin. Indian J Clin Biochem. 2007;22(Suppl):355.
- [Google Scholar]
- Phylogeny of mitochondrial DNA macrohaplogroup N in India, based on complete sequencing: implications for the peopling of South Asia. Am J Hum Genet. 2004;75:966-78.
- [Google Scholar]
- The dazzling array of basal branches in the mtDNA macrohaplogroup M from India as inferred from complete genomes. Mol Biol Evol. 2006;23:683-90.
- [Google Scholar]
- Updating phylogeny of mitochondrial DNA macrohaplogroup m in India: dispersal of modern human in South Asian corridor. PLoS One. 2009;4:e7447.
- [Google Scholar]
- Reanalysis and revision of the Cambridge reference sequence for human mitochondrial DNA. Nat Genet. 1999;23:147.
- [Google Scholar]
- Ancient mtDNA sequences in the human nuclear genome: a potential source of errors in identifying pathogenic mutations. Proc Natl Acad Sci USA. 1997;94:14900-5.
- [Google Scholar]
- A novel mitochondrial DNA-like sequence in the human nuclear genome. Genomics. 1999;60:67-77.
- [Google Scholar]
- Origin and expansion of haplogroup H, the dominant human mitochondrial DNA lineage in West Eurasia: the Near Eastern and Caucasian perspective. Mol Biol Evol. 2007;24:436-48.
- [Google Scholar]