Translate this page into:
Hospital-acquired infections due to carbapenem-resistant Providencia stuartii
For correspondence: Dr Tuhina Banerjee, Department of Microbiology, Institute of Medical Sciences, Banaras Hindu University, Varanasi 221 005, Uttar Pradesh, India e-mail: drtuhina@yahoo.com
-
Received: ,
This article was originally published by Wolters Kluwer - Medknow and was migrated to Scientific Scholar after the change of Publisher.
Abstract
Background & objectives:
During the course of a retrospective survey on healthcare associated infections (HAIs) due to carbapenem-resistant organisms, an unusual prevalence of HAIs due to carbapenem-resistant Providencia stuartii (CRPS) was found. Hence this study aimed to conduct the occurrence of P. stuartii associated HAIs with special reference to the drug resistance profiling of these isolates.
Methods:
Of the eight total HAI cases (7.5% of total HAIs and 33.3% of HAIs due to Enterobacterales) of CRPS infections included in this study, three were reported from ventilator-associated pneumonia (VAP), three were surgical site infections (SSIs), one was a catheter-associated urinary tract infection (CAUTI) and one was a bloodstream infection. All the eight CRPS isolates were tested for extended-spectrum β-lactamases production, AmpC hyperproduction as well as carbapenem resistance. Typing of the isolates was performed by repetitive extragenic palindromic polymerase chain reaction (REP-PCR).
Results:
All the eight isolates of CRPS were found to be AmpC hyperproducers, carbapenemase producers, and harboured chromosomally located blaNDM in seven isolates and blaIMP genes in one. All the cases with CRPS infections had prior history of colistin therapy along with prolonged hospital stay (>20 days). The cases were located in five different wards/intensive care unit (ICU) within the hospital in one year. However, strain typing by REP-PCR revealed 100 per cent similarity and clonal relatedness in all the seven isolates carrying blaNDM genes. Interestingly, routine hospital surveillance revealed a high carriage of P. stuartii in the axilla of patients admitted to the ICU.
Interpretation & conclusions:
The study findings suggest CRPS as an important cause of HAIs. This organism often goes unnoticed due to the burden of carbapenem resistance in other Enterobacterales and non-fermenters.
Keywords
AmpC
blaNDM
carbapenem-resistant P. stuartii
colistin
intensive care unit
There has been a recent rise in infections due to carbapenem-resistant Providencia stuartii (CRPS, P. stuartii) throughout the globe1. While the widespread occurrence of extended-spectrum β-lactamases (ESBL)-producing P. stuartii in hospitalized patients was realized much earlier2, outbreaks of CRPS in intensive care units (ICUs), followed by dissemination of such isolates in tertiary care hospitals have been gradually emerging3,4. Among other species of Providencia, P. stuartii is the most resistant species towards majority of the antimicrobial agents (AMAs)1. Once an infrequent cause of healthcare-associated infections (HAIs), this organism is being increasingly isolated from blood, respiratory samples and device related infections, especially in long-term care facilities5. Treatment of such infections is challenging owing to their intrinsic resistance to colistin and tigecycline1. Interestingly, this attribute has facilitated the emergence and spread of this organism as evident by studies indicating the association of its increasing prevalence with increased use of colistin in hospitals6. As developing countries are often the biggest producers and consumers of these classes of AMAs, P. stuartii could be an underrecognized threat in such setups due to the paucity of reports on this organism. Realizing the need, this study aimed to assess the occurrence of P. stuartii-associated HAIs in a tertiary care hospital in North India.
Material & Methods
The present study was conducted in the department of Microbiology and the associated 1500-bedded tertiary care hospital of Banaras Hindu University, Varanasi, in North India. In the course of a retrospective survey (between March, 2019 - February, 2020) on HAIs due to carbapenem-resistant Acinetobacter spp., a total of eight cases of HAI due to P. stuartii, constituting 7.5 per cent of all HAIs and 33.3 per cent of HAIs due to carbapenem-resistant Enterobacterales were detected. The study was approved by the Institutional Ethics Committee.
Isolates were revived from stock cultures that were maintained in cryovials containing 10 per cent glycerol at −80°C. The cultures were revived by gently scraping the surfaces of the frozen vials without thawing and plating a loopful on Luria Bertani agar, followed by overnight incubation. The revived isolates were further analyzed as mentioned below. Medical records were retrospectively reviewed for these cases.
The identification of the bacterial isolates was confirmed in the M50 Phoenix system (Becton Dickinson Microbiology Systems, Sparks, MD, USA). Antimicrobial susceptibility testing against amikacin (30 μg), gentamicin (10 μg), imipenem (10 μg), meropenem (10 μg), cefotaxime (30 μg), cefepime (30 μg), ampicillin (10 μg), amoxicillin–clavulanate (20/10 μg), piperacillin–tazobactam (100/10 μg), trimethoprim–sulfamethoxazole (1.25/23.75 μg), chloramphenicol (30 μg), ciprofloxacin (5 μg) and levofloxacin (5 μg) was done by disc diffusion method and corroborated with the results of BD Phoenix for the majority of the antimicrobials. For quality control, Escherichia coli ATCC 25922 was used. However, for imipenem, meropenem and minocycline, minimum inhibitory concentration (MIC) was detected by broth microdilution method as per standard protocols7. All the isolates were screened for probable ESBL production based on the revised zone diameters considering E. coli ATCC 25922 as control7. Detection of ESBL genes was done by multiplex PCR using primers for blaTEM, blaSHV, blaCTXM-1, 2,9 and blaOXA-2,10 as described elsewhere8. Screening for AmpC hyperproduction was done by cefoxitin disc (30 μg) and confirmed by disc antagonism test using cefoxitin (30 μg) and cefotaxime (30 μg) discs9. E. coli ATCC 25922 served as a control.
Carbapenemase production in the isolates was detected by Carba NP test (bioMerieux Diagnostics, France). Molecular detection of Class A and Class B carbapenemases was done by multiplex PCR as described per reference10,11. Previously characterized and published isolates were considered as positive controls for both ESBL and carbapenemase genes12. Molecular grade water was used as a negative control. All the primers used in the study are tabulated in Supplementary Table.
| Primer pairs | Sequence (5’- 3’) | Target | Base-pair | Reference |
|---|---|---|---|---|
| TEM F | ATGAGTATTCAACATTTCCG | blaTEM | 867 | 8 |
| TEM R | CTGACAGTT ACCAATGCTTA | |||
| SHV F | AGGATTGACTGCCTTTTTG | blaSHV | 392 | |
| SHV R | ATTTGCTGATTTCGCTCG | |||
| CTX-M-A | CGCTTTGCGATGTGCAG | blaCTXM-1,2,9 | 550 | |
| CTX-M-B | ACCGCGATATCGTTGGT | |||
| OXA I F | TCAACAAATCGCCAGAGAAG | blaOXA-10 | 276 | |
| OXA I R | TCCCACACCAGAAAAACCAG | |||
| OXA II F | AAGAAACGCTACTCGCCTGC | blaOXA-2 | 478 | |
| OXA II R | CCACTCAACCCATC CTACCC | |||
| GES-F | GCTTCATTCACGCACTATT | blaGES1-9, 11-20 | 323 | 10 |
| GES-MR | CGATGCTAGAAACCGCTC | |||
| IMI (NMC)-F1 | TGCGGTCGATTGGAGATAAA | blaIMI13 and | 399 | |
| IMI (NMC)-R1 | CGATTCTTGAAGCTTCTGCG | blaNMC-A | ||
| SME-F1 SME-R1 | ACTTTGATGGGAGGATTGGC ACGAATTCGAGCATCACCAG | blaSME1-3 | 551 | |
| KPCF2 | GTATCGCCGTCTAGTTCTGC | blaKPC2-13 | 638 | |
| KPCFR | GGTCGTGTTTCCCTTTAGCC | |||
| IMP-F | GGAATAGAGTGGCTTAAYTCTC | blaIMP | 232 | 11 |
| IMP-R | GGTTTAAYAAAACAACCACC | |||
| VIM-F | GATGGTGTTTGGTCGCATA | blaVIM | 390 | |
| VIM-R | CGAATGCGCAGCACCAG | |||
| NDM-F | GGTTTGGCGATCTGGTTTTC | blaNDM | 621 | |
| NDM-R | CGGAATGGCTCATCACGATC | |||
| REP1 | IIIGCGCCGICATCAGGC | - | - | 13 |
| REP2 | ACGTCTTATCAGGCCTAC |
Isolation of plasmid DNA was done using QIA Spin Mini Prep kit (Qiagen) based on alkaline lysis of bacterial cells. Detection of carbapenemase genes was repeated from the DNA of the isolated plasmids.
Strain typing was done by repetitive extragenic palindromic-PCR (REP-PCR). REP1- and REP2-specific primers were used for amplification as previously described13. Gel imaging was done using Bio-rad gel imager (BioRad Laboratories Pvt. Ltd, India) and dendrogram was constructed using NTSYS pc 2.02 software (NY, USA).
Results
In this study, the eight isolates of P. stuartii from different HAIs constituted 33.3 per cent of all HAIs due to carbapenem-resistant Enterobacterales, being the second-most common isolate after Klebsiella pneumoniae. The associated HAIs were from (VAP, 3 cases), (SSIs, 3 cases), (CAUTI, 1 case) and (BSIs, 1 case). These cases were located in five different locations including surgery, medicine, chest, urology wards and ICU of the hospital as shown in Table I. All the patients had prior colistin and/or tigecycline therapy and prolonged hospitalization of more than 20 days. The details of the cases are summarized in Table I.
| Characteristics | Cases (n) | |||||||
|---|---|---|---|---|---|---|---|---|
| C1 | C2 | C3 | C4 | C5 | C6 | C7 | C8 | |
| Sex/age (year) | 20/male | 28/male | 42/male | 30/male | 32/male | 57/male | 25/male | 18/male |
| Diagnosis | VAP | VAP | SSI | SSI | SSI | CAUTI | BSI | VAP |
| Hospital admission >20 days | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes |
| Admission site | ICU | ICU | Surgery ward | Surgery ward | Surgery ward | Urology ward | Medicine ward | Chest ward |
| Specimen | ETA | ETA | Pus | Pus | Pus | Urine | Blood | Sputum |
| P. stuartii isolation date | May 26, 2017 | September 12, 2017 | November 7, 2017 | November 22, 2017 | December 18, 2017 | December 21, 2017 | March 13, 2018 | April 24, 2018 |
| Outcome | Expired | Expired | Survived | Expired | Expired | Survived | Expired | Survived |
| Prior use of colistin | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes |
VAP, ventilator-associated pneumonia; SSI, surgical site infection; CAUTI, catheter-associated urinary tract infection; BSI, blood stream infections; ICU, intensive care unit; ETA, endotracheal aspirate; P. stuartii, Providencia stuartii
All the isolates were fully resistant (100%) to ampicillin, cefazolin, cefoxitin, ceftazidime, cefotaxime, cefepime, amoxicillin–clavulanate, piperacillin–tazobactam, imipenem, meropenem, amikacin, gentamicin, ciprofloxacin and levofloxacin, but were 100 per cent susceptible to aztreonam and minocycline, as shown in Table II. None of the targeted ESBL genes were detected. All the isolates were AmpC hyperproducers. All the isolates showed carbapenemase production by Carba NP test. Among the isolates, seven showed the presence of blaNDM-1 gene, while one was positive for blaIMP (Fig. 1). None of the carbapenemases were detected in the plasmids of the isolates. Strain typing revealed two different REP-PCR profiles. All the isolates harbouring blaNDM-1 were clonally related with 100 per cent similarity (Fig. 2).
| Isolate number | MIC (μg/ml) | ESBL gene | AmpC | Carbapenemase genes | |||||
|---|---|---|---|---|---|---|---|---|---|
| Doripenem | Meropenem | Imipenem | Aztreonam* | Minocycline | Class A | Class B | |||
| C1 | 8 | 8 | 128 | ≤2 | 2 | − | + | − | blaNDM |
| C2 | 32 | 64 | 128 | ≤2 | 2 | − | + | − | blaIMP |
| C3 | 32 | 32 | 128 | ≤2 | 2 | − | + | − | blaNDM |
| C4 | 32 | 8 | 128 | ≤2 | 2 | − | + | − | blaNDM |
| C5 | 4 | 8 | 256 | ≤2 | 2 | − | + | − | blaNDM |
| C6 | 4 | 8 | 256 | ≤2 | 2 | − | + | − | blaNDM |
| C7 | 4 | 8 | 128 | ≤2 | 2 | − | + | − | blaNDM |
| C8 | 4 | 8 | 128 | ≤2 | 2 | − | + | − | blaNDM |
*BD Phoenix M50 Antimicrobial Susceptibility Testing result. MIC, minimum inhibitory concentration; ESBL, extended-spectrum β-lactamases
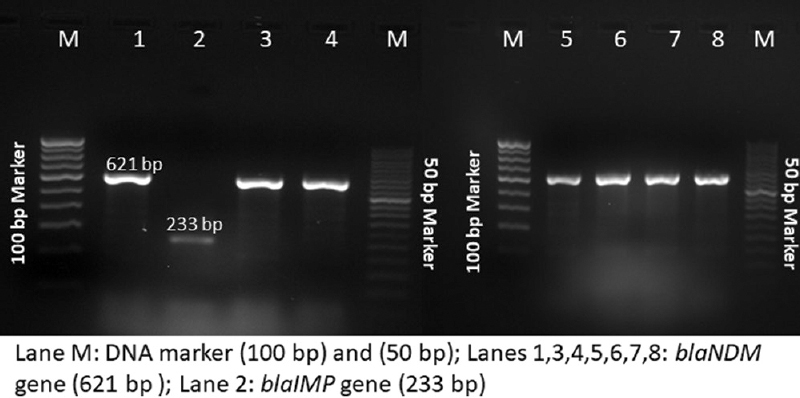
- Gel image showing the presence of Class B carbapenemases in the CRPS isolates. CRPS, carbapenem-resistant Providencia stuartii.

- Gel image of REP PCR in CRPS isolates showing similarity. REP PCR, repetitive extragenic palindromic polymerase chain reaction; CRPS, carbapenem-resistant Providencia stuartii.
Discussion
This study identified the emergence of CRPS-associated HAIs in a tertiary care hospital of India which is important because, to the best of our knowledge, this is the first extensive prevalence-based report on CRPS from the Indian subcontinent. There have been only a few reports on P. stuartii in global literature5. A quick literature search on the available studies reveals considerable dearth of data on P. stuartii from clinical cases. There have been infrequent reports on carbapenem-resistant P. rettgeri from other Asian countries including India, China, Pakistan, Nepal, Japan and Afghanistan. Only single cases have been reported along with other Enterobacterales5. The clinical relevance of P. stuartii as a nosocomial pathogen has not been studied or documented so far from these regions. This study too was the result of incidental findings of the emergence of this organism while trying to estimate the prevalence of carbapenem-resistant Acinetobacter spp.-associated HAIs. However, a handful of studies across the globe had already alerted about the emergence of P. stuartii as a potential nosocomial pathogen14-16.
The emergence of ESBL-producing P. stuartii was reported in a study from Italy in 20042 followed by reports of outbreaks from across the globe. Among the ESBLs, reports of association of certain genes like blaPER-1 have been increasingly associated with Providencia infections17. But in recent reports, P. stuartii-producing Class A and D ESBLs have been mentioned18. In this study, however, none of the tested ESBL genes were detected in the isolates. Instead, all the isolates were hyperproducers of AmpC beta-lactamases. Consequently, the isolates showed extensive resistance to almost all generations of cephalosporins and other penicillins along with beta-lactamase inhibitor combinations.
The presence of carbapenemase-encoding genes such as blaNDM-1 and blaIMP accounted for the carbapenem resistance phenotype of these isolates. Among the carbapenemase genes in P. stuartii, blaNDM-1 is reported to be the most common mechanism of carbapenem resistance5. However, carbapenem resistance in Providencia due to non-carbapenemase mechanisms has also been reported, where AmpC hyperproduction has accounted for carbapenem resistance3. In this type of non-carbapenemase mechanisms, the carbapenem-resistant phenotype is often due to de-repression of chromosomal AmpC production along with other alterations in outer membrane proteins, efflux pumps and penicillin-binding proteins. Both these mechanisms were seen in these isolates, a fact accounting for the considerably higher MIC for imipenem depicted in these isolates. The spread of these AmpC hyperproducing isolates contributing to carbapenem resistance is really bothersome owing to the limited therapeutic options available against these strains. Therapy with aztreonam and minocycline based on susceptibilities was the only therapeutic option in these infections. However, minocycline therapy is not a feasible option, especially in resource-limited countries like ours due to cost factors associated with minocycline therapy. Similarly, in the absence of effective agents for combination therapy with aztreonam in these cases, this option could not be utilized in the treatment of the patients. Consequently, failure to treat these cases with the appropriate choice of AMAs was a major challenge.
The clonal relatedness of these isolates hinted towards the hospital-wide dissemination of the same clone from one ward to the other. Although not within the scope of this study, we propose that frequent movement and regular duty exchanges of healthcare workers across these wards could be contributing towards the wide dissemination of these isolates through unsatisfactory infection control practices. It has been reported that multiple strains of P. stuartii, can be simultaneously present in a particular setting18. Studies have also suggested the ability of inter-hospital transfer in such well adapted hospital clones14.
Studies have predicted that excessive colistin usage in hospitals provides a survival advantage to these intrinsically colistin-resistant organisms, P. stuartii6. With the growing burden of carbapenem-resistant Enterobacterales and non-fermenters in the hospitals, colistin therapy as the last resort treatment is already on the rise. All the cases in the present study had received colistin therapy before the isolation of this organism. Similarly, it could be predicted that owing to the previously studied burden of carbapenem resistance in the study setup19, colistin usage was relatively high.
P. stuartii has been a recognized colonizer in long-term care facilities. Outbreaks of P. stuartii have often been controlled by emphasizing decontamination of common equipment or point prevalence survey for the detection of colonization20. The tertiary care hospital in this study had previously witnessed endemicity of Acinetobacter, especially in the ICUs19. To trace the possible reservoirs, recently, a surveillance protocol was planned and performed (December 2019 - February 2020) to screen the hospital environment including the high-touch surfaces such as bedrails, beddings, drip stands, suction tubes and skin surfaces of the patients21. Interestingly, the surveillance revealed the frequent presence of P. stuartii (32/90, 35.6%) in the axillary swabs of the patients. In addition, the presence of P. stuartii on the skin was significantly associated with those patients who were on colistin or polymyxin B therapy. Therefore, further studies should focus on whether skin colonization is the potential reservoir of P. stuartii for these HAIs.
The study was not without limitations. It was a retrospective survey, and the current trend of similar infections could not be assessed. Genome analysis of these isolates and study of other mechanisms of carbapenem resistance was not within the scope of this study. However, this is the first extensive report on the emergence of CRPS from one of the developing countries, which might be of significance to raise awareness on P. stuartii in HAIs and the real challenge of treating these infections. Therefore, early interventions in the form of curbing excessive usage of antibiotics, particularly colistin and prudent use of carbapenems to reduce selection of carbapenem-resistant organisms, should be endeavoured.
Further studies should be intended to estimate the actual burden of the problem and for early identification of this pathogen.
Financial support and sponsorship
None.
Conflicts of interest
This work was partly presented at the Annual conference of Indian Association of Medical Microbiologists in 2020 and published as a part of conference proceedings in the Ind J Med Microbiol (https://doi.org/10.1016/j.ijmmb.2021.08.281).
Acknowledgment:
The authors acknowledge Becton Dickinson India Pvt. Ltd., for providing consumables for this work. The authors also thank Banaras Hindu University for providing the basic infrastructure for carrying out the work.
References
- Clinical and drug resistance characteristics of Providencia stuartii infections in 76 patients. J Int Med Res. 2020;48:300060520962296.
- [Google Scholar]
- ESBL-producing multidrug-resistant Providencia stuartii infections in a university hospital. J Antimicrob Chemother. 2004;53:277-82.
- [Google Scholar]
- Outbreak of carbapenem-resistant Providencia stuartii in an Intensive Care Unit. Infect Control Hosp Epidemiol. 2012;33:627-30.
- [Google Scholar]
- Emergence of a pandrug-resistant VIM-1-producing Providencia stuartii clonal strain causing an outbreak in a Greek Intensive Care Unit. Int J Antimicrob Agents. 2015;45:533-6.
- [Google Scholar]
- First literature review of carbapenem-resistant Providencia . New Microbes New Infect. 2018;25:16-23.
- [Google Scholar]
- Growing prevalence of Providencia stuartii associated with the increased usage of colistin at a tertiary health care center. Int J Infect Dis. 2012;16:e646-646.
- [Google Scholar]
- Performance standards for antimicrobial susceptibility testingCLSI supplement M100 (29th ed). Wayne, PA, USA: CLSI; 2019.
- Long-term outbreak of Klebsiella pneumoniae &third generation cephalosporin use in a neonatal Intensive Care Unit in north India. Indian J Med Res. 2016;144:622-9.
- [Google Scholar]
- Presence of different beta-lactamase classes among clinical isolates of Pseudomonas aeruginosa expressing AmpC beta-lactamase enzyme. J Infect Dev Ctries. 2010;4:239-42.
- [Google Scholar]
- Multiplex PCR for rapid detection of genes encoding class A carbapenemases. Ann Lab Med. 2012;32:359-61.
- [Google Scholar]
- Multiplex PCR for detection of acquired carbapenemase genes. Diagn Microbiol Infect Dis. 2011;70:119-23.
- [Google Scholar]
- Extensively drug-resistant hypervirulent Klebsiella pneumoniae from a series of neonatal sepsis in a tertiary care hospital, India. Front Med (Lausanne). 2021;8:645955.
- [Google Scholar]
- Utility of whole-genome sequencing in characterizing acinetobacter epidemiology and analyzing hospital outbreaks. J Clin Microbiol. 2016;54:593-612.
- [Google Scholar]
- Dissemination of NDM-1 carbapenemase-producer Providencia stuartii strains in Romanian hospitals: A multicentre study. J Hosp Infect. 2019;103:165-9.
- [Google Scholar]
- Emergence of Carbapenem-Resistant Providencia rettgeri and Providencia stuartii Producing IMP-Type Metallo-β-Lactamase in Japan. Antimicrob Agents Chemother. 2020;64:e00382-00382.
- [Google Scholar]
- Case report:Providencia stuartii conjunctivitis. J Ophthal Inflamm Infect. 2016;29:1-3.
- [Google Scholar]
- Detection of PEr 1 extended-spectrum β-lactamase among nosocomial Providencia stuartii isolates in Tunisia. Tunis Med. 2014;92:258-61.
- [Google Scholar]
- Providencia species. Microbes. Available from: http://www.antimicrobe.org/b227.asp
- High prevalence and endemicity of multidrug resistant Acinetobacter spp in Intensive Care Unit of a tertiary care hospital, Varanasi, India. J Pathog. 2018;2018:9129083.
- [Google Scholar]
- Outbreak of carbapenem-resistant Providencia rettgeri in a tertiary hospital. S Afr Med J. 2016;107:31-3.
- [Google Scholar]
- Reservoir of Carbapenem-Resistant Acinetobacter baumannii in the hospital environment and colonization pressure: A surveillance-based study in Indian intensive care unit. Microb Drug Resist. 2022;28:1079-86.
- [Google Scholar]






