Translate this page into:
High prevalence of multi-drug resistant organisms in the gut flora of healthy rural individuals in northern India
For correspondence: Dr Saurabh Kedia, Department of Gastroenterology, All India Institute of Medical Sciences, New Delhi 110 029, India e-mail: dr.saurabhkedia@yahoo.com
-
Received: ,
Abstract
Background & objectives
Presence of resistant gut flora in the community is associated with increasing multi-drug resistance (MDR) infections. In this study, the prevalence of MDR organisms in the gut flora of a healthy rural population residing in northern India was determined.
Methods
Healthy individuals aged 18-45 yr from Nuh district, Haryana, India were included in this study. Risk factors associated with dysbiosis, diet, lifestyle and exposure to animals was assessed. Qualitative food frequency questionnaire and inflammatory diet score was calculated. Pathogens in stool sample were detected by MALDI-TOF. Evaluation of antimicrobial susceptibility was done by automated Vitek-2 System. The presence of antimicrobial resistance (AMR) genes was evaluated using PCR. An isolate having resistance to at least one antibiotic out of the three or more classes of antibiotics tested was labelled as MDR.
Results
Among 153 individuals included in this study (mean age-32.5±8.6 yr, females-58.2%, vegetarian-68.6%), the most frequent organism isolated was E. coli (n=137, 89.5%) followed by K. pneumoniae (n=19, 12.4%) and Enterobacter species (n=23, 15%). Forty seven (30.7%) individuals had sensitive and 42 (27.4%) had MDR organisms. Fifty one (33.3%) were positive for ESBL, 5 (3.3%) were positive for carbapenems, and 18 (11.8%) were positive for both genes. Age, gender, body mass index, diet pattern, or diet score were similar between participants with sensitive and resistant organisms. Resistance against fluoroquinolones was highest [92(48.7%)] among all isolates. Forty nine (25.9%), 25 (13.2%), 24 (12.7%) and 21 (11.1%) isolates, respectively were positive for blaTEM, blaSHV, blaCTXM-1 and OXA-48 genes.
Interpretation & conclusions
Overall the study findings suggest that 27 per cent individuals from rural northern India carry MDR organisms in their fecal flora, with an ESBL carriage rate of 44 per cent.
Keywords
Rural India
MDR Gram negative bacteria
gut microflora
Antimicrobial resistance (AMR) is a major public health problem due to infections with multi-drug resistant (MDR) organisms1. Irrational antibiotic use is responsible for increasing MDR infections. According to a recent report, India has some of the highest antibiotic resistance rates among bacteria that commonly cause infections in the community and healthcare facilities2. There is rising resistance to β-Lactams; several studies have reported high prevalence of cephalosporinases, extended spectrum β-lactamases (ESBLs), Amp C and Carbapenemase, amongst the common pathogens3. ESBL producing Enterobacterales are now included under the threat pathogen list of Center for disease control (CDC)4. Further, community acquired infection with ESBL producing E. coli is also quite frequent5.
Intestinal colonisationwith MDR organisms (MDROs) contributes to the AMR gene reservoir within the gut6. This concept of a ‘resistome’ encompasses the AMR gene profile of the entire resident gut microbiota. Though there are several studies evaluating the burden of resistome in hospitalized individuals and healthy individuals in the community from other parts of globe7, there is limited information on the prevalence of AMR genes in healthy individuals from India. The literature on this aspect from India is limited to a single community-based study, and two other studies in healthy neonates and children8-10.
A balanced community of inhabitant gut microbiota are crucial for human health and hence have been studied extensively in Indian as well as global populations11,12. Alteration in healthy gut flora, or dysbiosis has been extensively studied in many gastrointestinal, metabolic and neurologic disorders. Correction of dysbiosis by fecal microbial transplantation (FMT) can be a promising tool for microbiome manipulation and could be an effective therapy for these disorders13. Though FMT is regarded as a safe procedure, in a recent alert by the American Gastroenterology Association, FMT from healthy donors to immunocompromised recipients has reportedly led to sepsis with ESBLs carrying E. coli in two patients, of whom one succumbed14.
These findings underscore the urgent need for in-depth studies on AMR pattern in the healthy Indian population. This can help us understand the role and transmission dynamics of AMR in communities and can also aid in the selection of healthy donors for enhancing the safety and efficacy of FMT. Therefore, the present study was undertaken to determine the prevalence of MDROs and ascertain the genotypic profile of fecal AMR bacterial isolates in the gut flora of healthy rural population residing in a rural district of Northern India.
Material & Methods
This study was conducted at the department of Gastroenterology in collaboration with the departments of Community Medicine and Laboratory Medicine, All India Institute of Medical Sciences, New Delhi, after obtaining approval from the Institute Ethics Committee. Access to the village households was conducted through an existing tripartite MoU with the State Health Society, Haryana. Before undertaking the actual survey a formal permission was obtained from the head of the requisite village. A written informed consent was obtained from all the study participants before administering study procedures such as questionnaire and collection of stool samples.
Sample collection
Samples were collected from healthy individuals in Nuh district, Haryana, in northern India. The district has a population of 1.1 million, with predominant rural inhabitation (89%). The biggest block Nuh with 70 villages was identified of which 15 villages were randomly selected (Fig. 1). The median population size (range) of the selected villages was 2,203 (661-6,597), with median (range) households per villages as 414 (117-1,024), respectively. Random lanes from center of village was approached for sample, the household members were briefed about the study.
- Number of samples collected from different villages.
Healthy adults aged 18-45 yr with no history of concurrent acute medical illness or hospitalization in past one year, symptoms pertaining to acute gastrointestinal (GI) disease, history of chronic illness, antibiotics/antifungals/antivirals/painkillers intake over the past three months, and other risk factors which could influence the gut microbiome were recruited (Supplementary Table I). A detailed questionnaire was administered (Supplementary Material). Stool samples were collected in sterile containers containing Copan E-swabs and were transported at 4℃ to the laboratory. The sample was processed for pathogen isolation, antimicrobial susceptibility testing (AST) and genetic profile of AMR genes.
Pathogen detection
All collected samples were cultured on blood agar, MacConkey agar (BD Dickinson, Biosciences, San Jose, CA), and Chrome ColAPSE (Himedia) screening agar plates and incubated for 16-18 hours at 37°C. After incubation, plates were inspected for type of growth. In case of pure growth, the identification of microbes was done by Matrix-Assisted-Laser-Desorption-ionization-Time-of-Flight (MALDI-TOF Biomerieux, Marcy-I’Etoile, France). In the case of multiple colonies, isolation was done as per standard microbiological techniques, followed by MALDI TOF. The confirmed isolates were then cryopreserved for further use.
Antimicrobial susceptibility testing
AST was done by automated AST Vitek 2 System (Biomerieux Marcy-I’Etoile, France) along with disc-diffusion testing and broth-microdilution-testing methods. MIC were defined as per CLSI guidelines15. The ESBL and carbapenemase production and colistin resistance was predicted by MIC of ceftazidime, meropenem and colistin respectively. MDR was defined as isolates having resistance to at least one antibiotic out of the three or more classes of antibiotics tested16.
Antimicrobial resistance genomics
AMR genes for colistin, ESBL and carbapenemase were detected from isolated microbes using PCR as detailed in Supplementary Material. The primer sequence and expected amplicon size for respective genes is shown in Supplementary Table II.
Diet analysis
During screening, diet over the past seven days from the date-of-screening was recorded with diet recall method as mentioned in Supplementary Material. The inflammatory component of the diet was assessed by classifying diet into pro-inflammatory (negative score) and anti-inflammatory (positive score) food groups as per the scores given in the Supplementary Table III17.
Statistical analysis
Categorical variables were presented as numbers (percentage) and continuous as mean±standard deviation or median (IQR). Categorical variables were compared with chi-square test. Continuous variables were compared with Student’s t test for two comparisons and ANOVA for more than two comparisons. Correlation between resistance profiles of different antibiotic classes was done using Spearman’s correlation co-efficient. Analyses were done with SPSS v26 (Stata Corp, College Station, TX).
Results
A total of 153 healthy volunteers were enrolled from 15 villages, the sample distribution is detailed in Figure 1. Mean age of participants was 32.5+8.6 yr with 58.2 per cent being females. Mean body mass index (BMI) was 22.2+4.8 kg/m2. Among these 105 (68.6%) participants were vegetarian and 48 (31.4%) members consumed both vegetarian and non-vegetarian (mixed) diet. About 62.1 per cent of the participants were exposed to cattle (Table I) and the mean diet score was 5.2+4.3.
| Mean Age yr (Mean+SD) | 32.5 + 8.6 |
| Gender (Females)* | 80 (58.2) |
| Diet Pattern* | |
|---|---|
| Vegetarian | 105 (68.6) |
| Mixed | 48 (31.4) |
| Body mass index (Kg/m2) (Mean+SD) | 22.2 + 4.8 |
| Cattle in house* | 95 (62.1) |
| Diet Score(Mean + SD) | 5.2 + 4.3 |
| Number of participants | 153 |
| Number of participants with single organisms | 123 (80.4) |
| Number of participants with 2 organisms | 24(15.7) |
| Number of participants with 3 organisms | 6 (3.9) |
| Types of organisms isolated (n=189) | |
| Escherichia coli* | 137 |
| Klebsiella pneumoniae* | 19 |
| Enterobacter cloacae* | 11 |
| Enterobacter faecium* | 9 |
| Enterobacter aerogenes* | 3 |
| Klebsiella oxytoca* | 2 |
| Citrobacter koseri* | 1 |
| Citrobacter Freundii* | 1 |
| Escherichia fergusonii* | 1 |
| Enterococcus gallinarum* | 1 |
| Pseudomonas aeruginosa* | 1 |
| Shigella flexneri* | 1 |
| Shigella group* | 1 |
| Shigella sonei* | 1 |
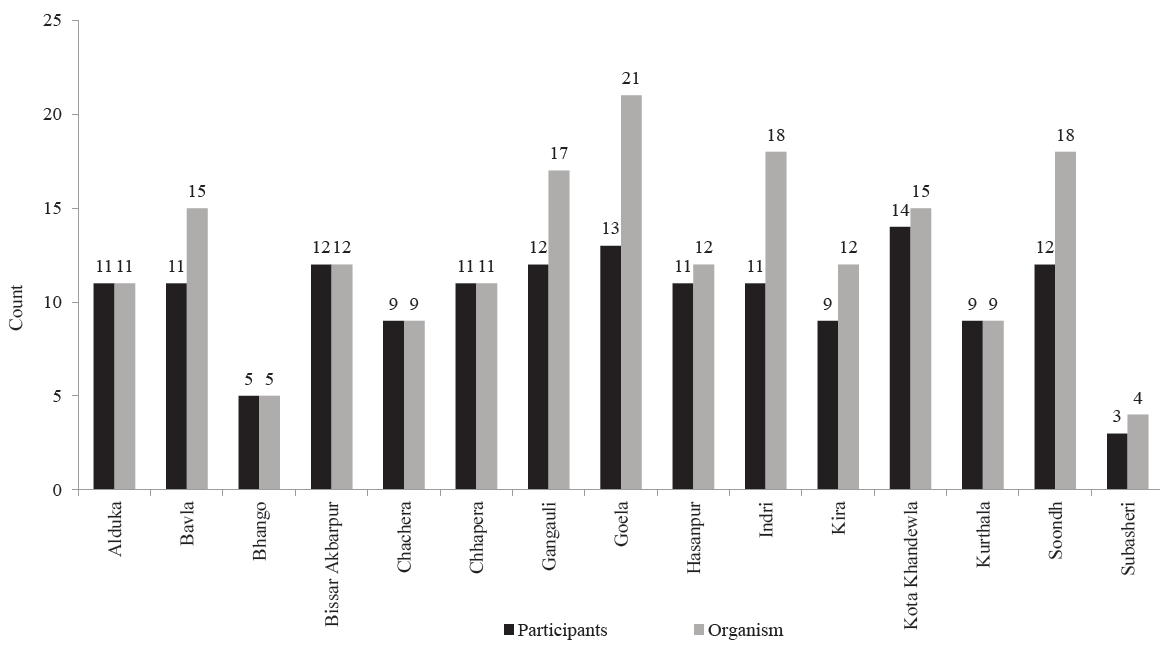
A total of 189 organisms were isolated from 153 participants, of these 123 (80.3%) participants were positive for only one organism, while the remaining were positive for >2 organisms (Table I). Village wise distribution of organisms is shown in Figure 2. Among 153 individuals, 137 had their culture positive for E. coli (89.5%). Among all organisms (n=189), the most frequent organism isolated was E. coli followed by K. pneumoniae and Enterobacter species Three participants who were positive for Shigella group bacteria were completely asymptomatic.

- Organisms isolated from different villages.
Antibiotic resistance profile of the study participants
Of all the participants, 24 (15.7%) had isolates that were sensitive to all drugs, without any AMR gene carriage. Phenotypically, 47 (30.7%) participants had isolates sensitive to all drugs, 64 (41.8%) had resistance to single/two drug classes, and 42 (27.4%) had MDR isolates. Isolates were resistant to a median of 3(IQR: 2-4) classes of drugs. ESBL genes were detected among isolates in 51 (33.3%) participants, only carbapenemases genes in 5(3.3%) and both genes in 18 (11.8%) participants. Among the E. coli, 28 (20.4%) were MDR isolates. ESBL genes were detected in 43(31.4%), carbapenemase gene in 5 (3.6%), and both genes in 15 (10.9%) isolates (Table II).
| Types of resistance | All Isolates (n=189) | All Participants (n=153) | Participants with E. coli (n=137) |
|---|---|---|---|
| Overall phenotypic resistance | 120 (63.5) | 106 (69.3) | 91 (66.4) |
| Phenotypic resistance profile in fecal samples | |||
| -No resistance | 69 (36.5) | 47 (30.7) | 46 (33.5) |
| -Single/ two drug resistance | 80 (42.3) | 64 (41.8) | 63 (45.9) |
| -MDR | 40 (21.1) | 42 (27.4) | 28 (20.4) |
| Genotypic resistance profile in fecal samples | |||
| -No AMR Gene | 100 (52.9) | 79 (51.6) | 74 (54.1) |
| -Only ESBL gene | 106 (32.8) | 51 (33.3) | 43 (31.1) |
| -Only carbapenemases gene | 6 (3.2) | 4 (2.6) | 5 (3.6) |
| -Both ESBL and carbapenemases genes | 21 (11.1) | 18 (11.1) | 14 (10.1) |
Data is presented as n(%). MDR, multi-drug resistant; AMR, antimicrobial resistance gene; ESBL, extended spectrum beta lactamase; E. coli, Escherichia coli
There was no difference in age, gender, BMI, diet pattern, presence of cattle in house or diet score between participants who had phenotypically sensitive organisms, those with single/two drug resistance, and those with MDR organisms (Table III).
| Parameters | Sensitive (n=47) | Single/ two drug resistant (n=64) | Multi-drug resistant (n=42) | P value |
|---|---|---|---|---|
| Age (yr) | 33.8 ± 9.2 | 31.9 ± 8.7 | 31.9 ± 7.5 | 0.43 |
| Gender, (females); n(%) | 27 (57.5%) | 35 (54.7%) | 27 (64.3%) | 0.61 |
| Body Mass Index | 22.2 ± 3.1 | 22.6 ± 4.9 | 22.3 ± 3.7 | 0.83 |
| Diet pattern; n(%) | 0.42 | |||
| Veg | 32 (68.1%) | 41 (64.1 %) | 32 (76.2%) | |
| Mixed | 15 (31.9%) | 23 (35.9%) | 10 (23.8%) | |
| Cattle in house; n(%) | 31 (66%) | 39 (60.9%) | 25 (59.5%) | 0.79 |
| Diet score | 5.3 ± 4.7 | 5.3 ± 4.1 | 4.9 ± 4.2 | 0.89 |
All isolates showed highest resistance against fluoroquinolones 92 (48.7%), followed by cotrimoxazole 56 (31.3%). Resistance pattern of isolates for other antibiotics is mentioned in Table IV. Among the E. coli also, highest resistance was present against fluoroquinolones 79 (57.2%), minocycline 25 (18.1%), aztreonam 6 (18.8%), and cephalosporin’s 15 (10.9%). Klebsiella sp. showed least resistance to fluroquinolones. All the isolates of Klebsiella were sensitive to carbapenems and only 10 per cent were resistant to third generation cephalosporins. Only Enterobacter sp. [2 (14.3%)] showed resistance to colistin. Citrobacter/Shigella revealed highest resistance to aminoglycosides [4 (66.7%)] (Table IV).
| Organisms | Colistin (n=169) | Beta-lactam + beta-lactamase inhibitor (n=179) | Third generation cephalon-sporin* (n=179) | Fourth generation cephalon-sporin* (n=179) | Aztreonam (n=178) | Carba-penem (n=179) | Amino-glycoside (n=179) | Fluro-quinolone (n=189) | Mino-cycline (n=178) | Tige-cycline (n=188) | Cotri-moxazole (n=179) |
|---|---|---|---|---|---|---|---|---|---|---|---|
| For all isolates | 2 (1.2) | 14 (7.8) | 18 (10.1) | 7 (3.9) | 30 (16.9) | 2 (1.1) | 10 (5.6) | 92 (48.7) | 31(17.4) | 2 (1.1) | 56 (31.3) |
| Per individual organisms | |||||||||||
| E. Coli (n=137) | 0 | 13 (9.4) | 15 (10.9) | 7 (5.0%) | 26 (18.8) | 2 (1.4) | 6 (4.3) | 79 (57.2) | 25 (18.1) | 1 (0.7) | 49 (35.5) |
| Enterococcus (n=10) | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 4 (40) | 0 | 0 | 0 |
| Enterobacter (n=14) | 2 (14.3) | 0 | 1 (7.1) | 0 | 0 | 0 | 0 | 3 (21.4) | 2 (14.3) | 0 | 3 (21.4) |
| Klebsiella (n=21) | 0 | 0 | 2 (9.5) | 0 | 3 (14.3) | 0 | 0 | 4 (19) | 2 (9.5) | 0 | 2 (9.5) |
| Pseudomonas/ Citrobacter/ Shigella (n=6) | 0 | 1 (16.7) | 0 | 0 | 1 (16.7) | 0 | 4 (66.7) | 2 (33.3) | 2 (33.3) | 1 (16.7) | 2 (33.3) |
| ESBL genes present | 1(0.5) | 4 (0.22) | 2 (0.11) | 2 (0.11) | 19 (10.6) | 1 (0.5) | 6 (0.33) | 30 (15.8) | 23 (12.9) | 1 (0.5) | 33 (18.4) |
| Carbapenemase genes present | 0 (0) | 3 (0.16) | 2 (0.11) | 3 (0.16) | 10 (5.6) | 1 (0.5) | 2 (0.11) | 10 (5.2) | 6 (3.1) | 0 (0) | 3 (1.6) |
Data is presented as n(%),
There was significant resistance correlation (or a trend towards correlation) between resistance to Beta Lactam/ Beta-lactamase inhibitor combination and all other classes of antibiotics, except colistin. Similarly, resistance to beta-lactam/beta-lactamase inhibitor combination, cephalosporins, aztreonam and carbapenems had significant correlation between each other (Supplementary Table IV).
Antimicrobial resistance gene profile
The proportion of ESBL (43.9%) gene was almost three times the proportion of carbapenemase gene (14.3%) among all isolates. Highest ESBL and carbapenemase positivity was seen for Klebsiella species (57.1 and 19%, respectively; Table V. Twenty eight out of 40 MDR isolates (70%), 29/80 (36.5%) single/two drug resistant isolates, and 26/69 (37.7%) phenotypically sensitive isolates, respectively, were ESBL positive. The genotypic resistance profile of all isolates and individual organisms is mentioned in Table V. None of the isolates were positive for Mcr1 and Mcr5 genes.
| Organisms | ESBL | Carbapenemase | |||||||
|---|---|---|---|---|---|---|---|---|---|
| TEM | SHV | CTXM | OXA-1 (n=180) | Overall ESBL* | KPC | NDM | OXA-48 | Overall carbapenemases* | |
| For all isolates | 49 (25.9) | 25 (13.2) | 24 (12.7) | 11 (6.1) | 83 (43.9) | 7 (3.7) | 1 (0.5) | 21 (11.1) | 27 (14.3) |
| Per individual organisms | |||||||||
| E. Coli (n=137) | 36(26.3) | 12 (8.7) | 21 (15.3) | 11 (8) | 72 (52.6) | 2 (1.5) | 1 (0.7) | 20 (14.6) | 21 (15.3) |
| Enterococcus (n=10) | 4 (40) | 0 | 0 | 0 | 4 (40) | 0 | 0 | 0 | 0 |
| Enterobacter (n=14) | 3 (21.4) | 1 (7.1) | 1 (7.1) | 0 | 5 (35.7) | 1 (7.1) | 0 | 0 | 1 (7.1) |
| K. pneumoniae/(n=21) | 4 (19) | 12 (57.1) | 1 (4.8) | 0 | 12 (57.1) | 4 (19) | 0 | 1 (4.8) | 4 (19) |
| Pseudomonas/Citrobacter/Shigella (n=6) | 2 (33.3) | 0 | 1 (16.7) | 0 | 3 (50) | 0 | 0 | 0 | 0 |
The phenotypic and genotypic profiling showed that ∼50 per cent of the isolates with resistance to various drugs carried ESBL genes: beta-lactam (64%), aztreonam (56.6%), aminoglycoside (40%) and minocycline (61.2%), while 16.3 to 50 per cent isolates carried carbapenemase gene Table IV.
Discussion
We report a high proportion of healthy individuals with fecal carriage of MDR organisms in this study. Healthy individuals from Nuh district were selected through an extensive questionnaire to rule out possible factors which could affect the AMR carriage, and only 31 per cent of the study participants had no fecal carriage for drug resistant organisms. Approximately 42 per cent individuals carried isolates with resistance against single/two drug classes, and 27 per cent carried MDRO. As per a Niti Aayog report18, Nuh is the most backward district in India with poor health and education infrastructure. We had previously shown that, among healthy individuals, the microbiome of rural individuals was healthier than urban12. Though we did not have a comparative group of urban healthy adults, the prevalence of MDR in rural individuals was more than expected. Use of antibiotics in deprived areas like Nuh is increasing, possibly due to the increasing rate of irrational antibiotics prescription by informal practitioners in these villages that might be contacted first for simple illnesses. Although, human antibiotic use is the most important factor, other factors such as animal to human transfer (due to indiscriminate antibiotic use in animals and food industry), effect of diet, presence of AMR genes in waste and effluents could also be responsible for increasing MDR burden in healthy individuals19.
In a similar study from Chandigarh, India, overall prevalence of antibiotic resistance in healthy gut flora was found to be 70.5 per cent, similar to the phenotypic resistance in this study (69.3%). However, the prevalence of MDR was significantly high in this study, 27 per cent, as compared to only 2.4 per cent in Chandigarh8, which could be ascribed to differences in population (rural vs. semi-urban), exposure of residents to livestock (62% in this study), and antibiotic use patterns. In other rural-based studies from central India, 70 per cent of the MDR was seen in fecal flora in children of 1-3 yr age10. Similar heterogeneity has also been observed in other studies, and prevalence of MDR positivity varied from 51 per cent to as high as 93 per cent9,10,20-23. Possible reasons for this heterogeneity could be the nature of participants, definition of MDR, method of detection, and similar reasons as described above. As an extension of the healthy gut microbiome study described above12, the AMR profiling of five dominant commensals revealed that all the dominant gut bacteria were MDR, being resistant to at least seven different antimicrobials20. Therefore, multi-drug resistance is also being reported in commensals, possibly due to their inadvertent exposure to antibiotics and toxic compounds.
The predictors of MDR organism carriage include higher socio-economic status, exposure to antibiotics or hospitalization over the past one year, and recent proton pump inhibitor and non-steroidal anti-inflammatory drug (NSAID) use21-23. We selected individuals from the same geography having a similar lifestyle to control for confounders which can affect the fecal resistome. Among the possible predictors which were measured in this study age, gender, diet, and BMI did not have any influence. Diet is one of the major determinants of gut microbiome24, although the effect of diet on gut resistome and AMR gene profile has been less explored25,26. Possible reasons for lack of association between diet and AMR profile could be rather uniform diet in the same geography, and bias associated with recall method of diet assessment.
Approximately 44 per cent isolates were ESBL gene positive, and 14 per cent were positive for Carbapenems gene, similar proportions being observed among E. coli. In a recent meta-analysis, globally, 21.1 per cent of inpatients in healthcare setting, and 17.6 per cent healthy individuals had the presence of ESBL E. coli in fecal samples7. However, the prevalence in healthy Southeast Asians in this meta-analysis was 35.3 per cent, similar to our study. In the study from Chandigarh, prevalence of ESBL genes among cephalosporin resistant isolates was 1.9-25 per cent (different genes)8. In the present study, among 18 cephalosporin resistant isolates, 10 were ESBL positive. In another study of 115 healthy volunteers, ESBL-producing E. coli had a prevalence of 19 per cent, with CTX-M being the predominant type followed by TEM, unlike our study, where TEM was most common followed by CTX-M27.
Among 40 MDR isolates, about 70 per cent were ESBL positive, while 1/3rd of phenotypically sensitive isolates also carried ESBL genes. This phenotypic-genotypic discordance could be explained by the presence of silent/cryptic or proto resistance genes, which can encode AMR but may not be expressed or may undergo modifications after expression19.
Similar to other prevalence studies, E. coli was the most common organism isolated followed by Klebsiella species. Overall E. coli carriage among individual participants was 89.5 per cent (137/153), similar to that reported by Martinson et al28, (⁓90%). Carriage rate for K. pneumoniae (12.4%), was also similar to the review by Podschum R et al29 (K. pneumonaie fecal carriage in Indian population: 5-38%). Surprisingly, Shigella was detected in three individuals, though all were asymptomatic. Asymptomatic Shigella carriage has also been reported in other studies30. Highest antibiotic resistance was seen for fluroquinolones followed by cotrimoxazole. Least resistant antibiotics were colistin, tigecycline and carbapenems. Like our study, 41.5 per cent fluoroquinolones resistance was seen in Chandigarh study, although unlike ours, maximum resistance was seen for cephalosporins. Carbapenem resistance was also similar between the two studies. Heterogeneity on this aspect has also been reported in other studies. In a study from rural Thailand, maximum resistance was seen against tetracycline, and fluoroquinolones resistance was seen in only 19.7 per cent21. Studies from Vietnam have reported high frequency of colistin resistance and mcr gene carriage, possibly due to significant colistin use in livestock31,32. In the present study, only two isolates were colistin resistant and none carried mcr gene.
Though the study was strengthened by detailed dietary evaluation and estimation of phenotypic and genotypic resistance profile, but reported predictors of MDR and ESBL carriage in stool like history of antibiotic use and hospitalization beyond three months was not obtained. However, the study sample was well characterized with the questionnaire and we ruled out maximum factors that could affect the gut microbiome in short-term. The sample size was reasonable enough to characterize the AMR profile but a multicentric large cohort study should be conducted in order to evaluate the effect of diet and lifestyle on gut microbiome. Also, with advancement in the techniques, gut resistome through metagenomics approach should also be evaluated in further studies. A more detailed demographic data on socioeconomic status would also help us to understand the spread of resistance in healthy population.
To conclude we report that 27 per cent individuals from rural northern India are positive for MDR organisms in their fecal flora, with an ESBL carriage rate of ∼40 per cent. These findings provide important insights on burden of MDR in the community, and a roadmap for urgent antibiotic stewardship measures to control the imminent threat of community associated MDR infections.
Acknowledgment
Authors acknowledge the technical support provided by the departments of Microbiology, Community Medicine and Gastroenterology, All India Institute of Medical Sciences, New Delhi, India.
Financial support & sponsorship
This study was funded by Intramural grant from the All India Institute of Medical Sciences (Project Code: AC-14) and partly by a grant from Indian Council of Medical Research (Project Code: I-1203).
Conflicts of Interest
None.
Use of artificial intelligence (AI)-assisted technology for manuscript preparation
The authors confirm that there was no use of AI-assisted technology for assisting in the writing of the manuscript and no images were manipulated using AI.
References
- Editorial commentary: the dawning of microbiome remediation for addressing antibiotic resistance. Clin Infect Dis. 2016;62:1487-8.
- [Google Scholar]
- Scoping report on antimicrobial resistance in India. November 2017. Available from: http://www.dbtindia.nic.in/wpcontent/uploads/ScopingreportonAntimicrobialresistanceinIndia.pdf, accessed on February 15, 2019.
- Trends in antibiotic resistance among major bacterial pathogens isolated from blood cultures tested at a large private laboratory network in India, 2008-2014. Int J Infect Dis. 2016;50:75-82.
- [Google Scholar]
- The biggest antibiotic-resistant threats in the U.S. Available from: https://www.cdc.gov/drugresistance/biggest-threats.html, accessed on March 15, 2024.
- Acquisition of multidrug-resistant enterobacterales during international travel: a systematic review of clinical and microbiological characteristics and meta-analyses of risk factors. Antimicrob Resist Infect Control. 2020;9:71.
- [Google Scholar]
- Intestinal microbiota and antibiotic resistance: perspectives and solutions. Hum Microbiome J. 2018;9:11-5.
- [Google Scholar]
- Comparison of the global prevalence and trend of human intestinal carriage of ESBL-producing Escherichia coli between healthcare and community settings: a systematic review and meta-analysis. JAC Antimicrob Resist. 2022;4:dlac048.
- [Google Scholar]
- Antibiotic-resistant Enterobacteriaceae in healthy gut flora: A report from north Indian semiurban community. Indian J Med Res. 2019;149:276-80.
- [Google Scholar]
- Community acquisition of β-lactamase producing Enterobacteriaceae in neonatal gut. BMC microbiol. 2013;13:1-6.
- [Google Scholar]
- Antibiotic resistance in an Indian rural community: a ‘One-Health’ observational study on commensal coliform from humans, animals, and water. Int J Environ Res Public Health. 2017;14:386.
- [Google Scholar]
- The healthy microbiome–what is the definition of a healthy gut microbiome? Gastroenterology. 2021;160:483-94.
- [Google Scholar]
- Analysis of the gut microbiome of rural and urban healthy Indians living in sea level and high-altitude areas. Sci rep. 2018;8:10104.
- [Google Scholar]
- European consensus conference on faecal microbiota transplantation in clinical practice. Gut. 2017;66:569-80.
- [Google Scholar]
- Important safety alert regarding use of fecal microbiota for transplantation and risk of serious adverse reactions due to transmission of multi-drug resistant organisms. Available from: https://www.fda.gov/vaccines-blood-biologics/safety-availability-biologics/important-safety-alert-regarding-use-fecal-microbiota-transplantation-and-risk-serious-adverse, accessed on March 15, 2024.
- Performance standards for antimicrobial susceptibility testing (27th ed). CLSI document M100Wayne PA; 2017.
- Multidrug-resistant, extensively drug-resistant and pan drug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin Microbiol Infect. 2012;18:268-81.
- [Google Scholar]
- Development and application of a plant-based diet scoring system for Japanese patients with inflammatory bowel disease. Perm J. 2016;20:16-019.
- [Google Scholar]
- Nuh at Bottom of Niti Aayog’s 101 Most Backward Districts. The Hindu 2018 Available from: https://www.thehindu.com, accessed on March 29, 2018
- [Google Scholar]
- The human gut resistome: Current concepts & future prospects. Indian J Med Res. 2019;150:345-58.
- [Google Scholar]
- Molecular insights into antimicrobial resistance traits of commensal human gut microbiota. Microbl Ecol. 2019;77:546-57.
- [Google Scholar]
- Prevalence of and risk factors associated with faecal carriage of CTX-M β-lactamase-producing Enterobacteriaceae in rural Thai communities. J Antimicrob chemother. 2012;67:1769-74.
- [Google Scholar]
- High prevalence and risk factors of fecal carriage of CTX-M type extended-spectrum beta-lactamase-producing Enterobacteriaceae from healthy rural residents of Taian, China. Front Microbiol. 2015;6:239.
- [Google Scholar]
- Gastrointestinal carriage of Klebsiella pneumoniae in a general adult population: a cross-sectional study of risk factors and bacterial genomic diversity. Gut Microbes. 2021;13:1939599.
- [Google Scholar]
- Effect of diet on the intestinal microbiota and its activity. Curr Opinion Gastroenterol. 2014;30:189-95.
- [Google Scholar]
- Impact of long-term dietary habits on the human gut resistome in the Dutch population. Sci Rep. 2022;12:1892.
- [Google Scholar]
- Early-life formula feeding is associated with infant gut microbiota alterations and an increased antibiotic resistance load. Am J Clin Nutr. 2022;115:407-21.
- [Google Scholar]
- Fecal carriage rates of extended-spectrum β-lactamase-producing Escherichia coli among antibiotic naive healthy human volunteers. Microb Drug Resist. 2015;21:59-64.
- [Google Scholar]
- Escherichia coli residency in the gut of healthy human adults. EcoSal Plus. 2020;9 10.1128/ecosalplus
- [Google Scholar]
- Klebsiella spp. as nosocomial pathogens: epidemiology, taxonomy, typing methods, and pathogenicity factors. Clin Microbiol Rev. 1998;11:589-603.
- [Google Scholar]
- Asymptomatic salmonella, Shigella and intestinal parasites among primary school children in the eastern province. J Family Community Med. 1995;2:36-40.
- [Google Scholar]
- Prevalence of antimicrobial resistance genes and integrons in commensal Gram-negative bacteria in a college community. Microb Drug Resist. 2020;26:1227-35.
- [Google Scholar]
- Prevalence of mcr-1 among cefotaxime-resistant commensal Escherichia coli in residents of Vietnam. Infect Drug Resist 2019:3317-25.
- [Google Scholar]






