Translate this page into:
Heterologous expression of porcine elongase 6 (ELOVL6) gene in a human cell line
*For correspondence: sujoylab.office@gmail.com
-
Received: ,
This is an open access article distributed under the terms of the Creative Commons Attribution-NonCommercial-ShareAlike 3.0 License, which allows others to remix, tweak, and build upon the work non-commercially, as long as the author is credited and the new creations are licensed under the identical terms.
This article was originally published by Medknow Publications & Media Pvt Ltd and was migrated to Scientific Scholar after the change of Publisher.
Sir,
Obesity, insulin resistance, diabetes and associated cardiovascular disorders are highly prevalent lifestyle diseases12. Several attempts have been made worldwide in finding effective remedies. These included targeting of various adipogenic enzymes3, enzymes regulating fatty acid metabolism, like, acetyl-coenzymeA carboxylase, fatty acid synthase and stearoyl-CoA desaturase14567 or inducing non-shivering thermogenesis to help bypass the caloric contribution8. Yet the complete cure remained elusive.
Metabolic syndromes were typically accompanied by dysregulated lipogenesis and steroidogenesis. Elongase of long chain fatty acids family 6 (Elongase6, ELOVL6) is a rate-limiting enzyme catalyzing the chain elongation of palmitate (C16:0) to stearate (C18:0) and palmitoleate (C16:1n-7) to vaccinate (C18:1n-7)39 and it controls tissue fatty acid composition. Therefore, this microsomal enzyme is found ubiquitously in almost every tissue that supports active lipogenesis and steroidogenesis10.
It was established that Elongase6-deficient mice became obese and developed hepatosteatosis when fed with a high-fat diet or when mated to leptin-deficient ob/ob mice1011, but these animals exhibited improved insulin resistance10. ELOVL6 overexpression was noted in rats under abnormal feeding schedule12. Thus, it would be imperative to create an overexpression model to help evaluate Elongase6 further as a potential therapeutic target for metabolic diseases.
Pig, a naturally obese animal with high body fat content but exhibiting no metabolic syndrome, was considered as most preferred biomedical model for applications in human13. Therefore, biological systems in both species should be compatible to each other. This was not thoroughly verified, except some limited reports, where pig was used for culturing transgenic organs and for transplantation14. To facilitate and understand the interspecies compatibility using a cellular model, we attempted here to overexpress porcine Elongase6 in a human cell line, HEK293T. Thus, the present study was aimed to make an expression cassette with porcine Elongase6 (ELOVL6) gene and also to ensure its constitutive expression in a heterologous host (human).
To clone the porcine ELOVL6 gene, adipose tissue precursor, mesenchymal stem cells (MSCs) were isolated from bone marrow of adult pig available at local slaughter house (Fig. 1A) following the previously established protocol15, and subsequently total RNA was isolated from MSCs. RNA was converted into cDNA and it was finally used as a template for PCR amplification of Elongase6 open reading frame (ORF).
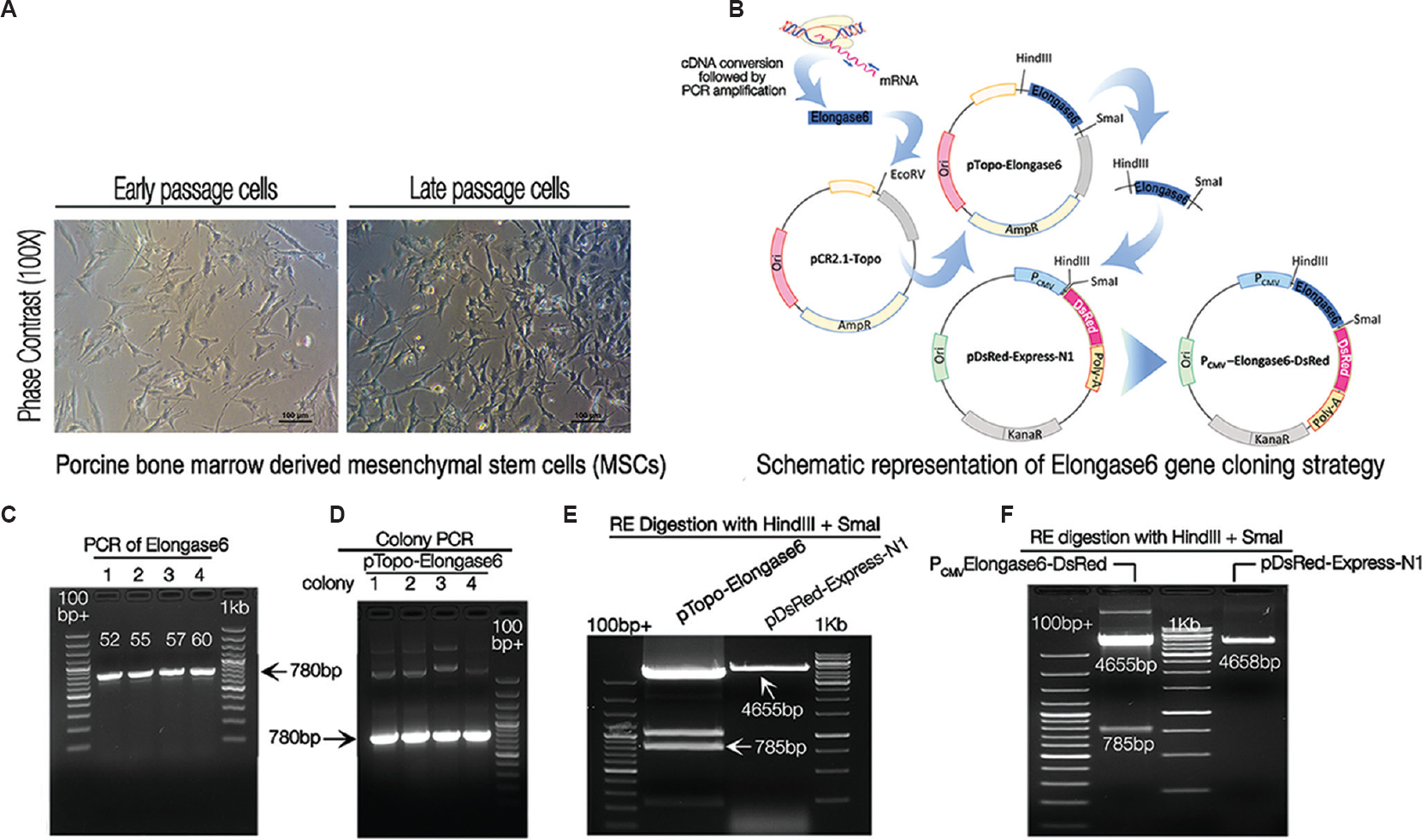
- Cloning and sequencing of porcine Elongase6 open reading frame (ORF). Bright field images of in vitro propagated bone marrow derived porcine mesenchymal stem cells, used as a source for cloning the gene. (A) The photomicrograph showed the characteristic morphology of porcine mesenchymal stem cells in monolayer during early and late passage, respectively. Scale bar=100μm (B) Schematic diagram showing stepwise Elongase6 open reading frame cloning. Elongase6 open reading frame was PCR amplified and cloned into a cloning vector pCR®2.1-TOPO® (designated as pTopo-Elongase6). Next, Elongase6 ORF was moved from cloning vector to expression vector pDsRed-Express-N1 (named the recombinant plasmid as PCMV-Elongase6-DsRed). (C) Gel picture showing PCR amplified 780bp Elongase6 open reading frame on 1 per cent agarose gel. Lanes 1-4 show PCR amplified bands at four different annealing temperatures namely, 52, 55, 57 and 60°C, respectively. (D) Positive clones were identified by colony PCR (lanes 1, 2, 3 and 4 different colonies). (E) Restriction enzyme digestion and gel cleaning of the insert (Elongase6 open reading frame) and backbone (pDsRed-Express-N1): For cloning of Elongase6 in an expression vector, pDsRed-Express-N1 and pTopo-Elongase6 were digested with HindIII and SmaI Res. DNA fragments of size 4655bp (backbone) and 785bp (Insert) were purified from 1 per cent agarose gel and used for further ligation reaction to generate expression vector (PCMV-Elongase6-DsRed). Lanes with samples were made wider (by removing gap between two adjacent combs with cello tape) for loading larger quantity of samples. (F) Screening of bacterial clones positive for PCMV-Elongase6-DsRed plasmid by restriction enzymes digestion: Plasmid was digested with HindIII and SmaI (empty pDsRed-Express-N1 vector as a negative control) and run on 1 per cent agarose gel. Gel picture confirmed the generation of recombinant plasmid, as restriction enzymes digestion released 785bp insert and 4655bp backbone fragments. 100bp+ denotes the GeneRuler™ 100bp Plus DNA Ladder (Fermentas), 1kb denotes StepUp™ 1 kb DNA Ladder (GeNei™).
In the present study, two separate gene constructs were developed, one for cloning of the gene and the other for its expression (Fig. 1B). For cloning, primers (forward 5’AAGCTTGCCACCATGAACATGTCAGTGTT GAC3’ and reverse 5’CCCGGGATCTTGCCGATG TAGGCCTCAAA G3’) were designed (using Design Tool https://www.ncbi.nlm.nih.gov/tools/primer-blast/) and synthesized (from IDT®, USA). Elongase6 ORF was PCR amplified using KOD FX (TOYOBO, Japan) polymerase in a 25 μl reaction containing 25 ng cDNA following the manufacturer's protocol with four annealing temperatures (52, 55, 57 and 60°C). PCR products were electrophoresed on 1 per cent agarose gel (Amresco®, USA) and 780bp DNA bands stained with ethidium bromide (Amresco®) (0.5 μg/ml) were visualized under ultraviolet transilluminator (ChemiDoc™ XRS+, Bio-Rad, USA) (Fig. 1C). These products were cloned in cloning vector pCR® 2.1-TOPO® (Invitrogen, USA) and the resultant recombinant vector was named as pTopo-Elongase6. Four recombinant clones were screened by colony PCR and it was found that all the four colonies were positive for Elongase6 insert, evident by the presence of 780bp specific bands (Fig. 1D). In addition, the fragment was sequenced and confirmed to be porcine Elongase6 gene (data submitted to GenBank, Accession No. KX237246).
To express the cloned fragment in mammalian cells, the Elongase6 ORF was moved from pTopo-Elongase6 (source plasmid) to pDsRed-Express-N1 (destination plasmid, Clontech, USA). Both the source and destination plasmids were digested with HindIII and SmaI restriction enzymes (RE) to release the desired insert and backbone fragments (785 and 4655bp, respectively) that were gel-purified (Fig. 1E). Following ligation and transformation, the antibiotic resistant recombinant colonies were produced.
One colony was selected for plasmid isolation and subsequently digested with HindIII and SmaI REs. The colony was found to be positive for recombinant plasmid PCMV-Elongase6-DsRed as it released the expected 785bp fragment (Fig. 1F) but digestion of empty vector pDsRed-Express-N1 expectedly yielded 4658bp linear fragment (Fig. 1E).
Porcine Elongase6 sequence data were subjected to comparative analysis (using BLAST tool at NCBI site). The analysis revealed that porcine Elongase6 shared 94 and 96 per cent of sequence identity with human Elongase6 at nucleotide and deduced amino acid level, respectively (Fig. 2). However, neither any amino acid residue potential to undergo glycosylation (http://www.cbs.dtu.dk/services/NetNGlyc/; http://comp.chem.nottingham.ac.uk/cgi-bin/glyco/bin/getparams.cgi) nor any B cell epitope (http://www.imtech.res.in/raghava/igpred/index.html; http://tools.iedb.org/bcell/; http://www.cbs.dtu.dk/services/BepiPred/) in porcine Elongase6 could be predicted.

- Comparative sequence analysis of Elongase6 gene in porcine and human: Porcine Elongase6 gene was cloned/sequenced (KX237246) and compared to human Elongase6 gene sequence (NM_24090.2) using BLAST tool. The similar analysis was performed with deduced amino acid sequences for proteins from both species. Results indicated that both species shared a high degree (94% and 96% of sequence identity) of relatedness for this gene.
In the construct, the ORF was put under strong cytomegalovirus immediate early (CMV-IE) promoter for its constitutive expression (Fig. 1B). The cloning primers were such designed that transgene Elongase6 expressed with DsRed as a single fusion protein. To check the functional activity of the PCMV-Elongase6-DsRed recombinant clone, in vitro expression study was carried out in HEK293T cells (National Centre for Cell Science, Pune). Before using the construct in heterologous HEK293T cell line, transfection experiments were conducted in cells from porcine origin (homologous system) and it was found that the construct could be delivered on an average to 20 per cent of cells. Once this information became available, HEK293T cells were cultured and transfected with PCMV-Elongase6-DsRed vector using polyethylenimine (PEI; Sigma, USA) following manufacturer's protocol. Simultaneous transfections with pDsRed-Express-N1 plasmid as vector control and sole PEI as mock control were also carried out. Forty eight hours post-transfection, cells of all the groups were fixed in 4 per cent paraformaldehyde (PFA), permeabilized with 0.2 per cent Triton X100, blocked with 2 per cent bovine serum albumin and immunostained with rabbit anti-Elongase6 specific primary antibody (Sigma, USA) (1:500 dilutions) followed by Alexa flour 488-labelled goat anti-rabbit IgG (Life Technologies™, USA, 1:1000 dilution) as secondary antibody. After nuclear staining with 4’,6-diamidino-2-phenylindole dihydrochloride (DAPI), cells were observed under fluorescence microscope.
As expected, the mock transfected cells (Fig. 3A1–5) showed only blue fluorescence (Fig. 3A2 and A5) for nuclear stain with DAPI. In vector control, (Fig. 3B1–5) cells transfected with pDsRed-Express-N1 displayed both red (Fig. 3B3 and B5) and blue (Fig. 3B2 and B5) fluorescence for functional DsRed protein along with the characteristic DAPI in nuclei. In the treatment group, (Fig. 3C1–5) cells transfected with PCMV-Elongase6-DsRed exhibited characteristic blue fluorescence (Fig. 3C2 and C5) for DAPI, red fluorescence (Fig. 3C3 and C5) for DsRed and green fluorescence (Fig. 3C4 and C5) for porcine Elongase6 protein. Therefore, it was evident that the gene construct was functional and produced Elongase6-DsRed fusion protein in HEK293T cells.
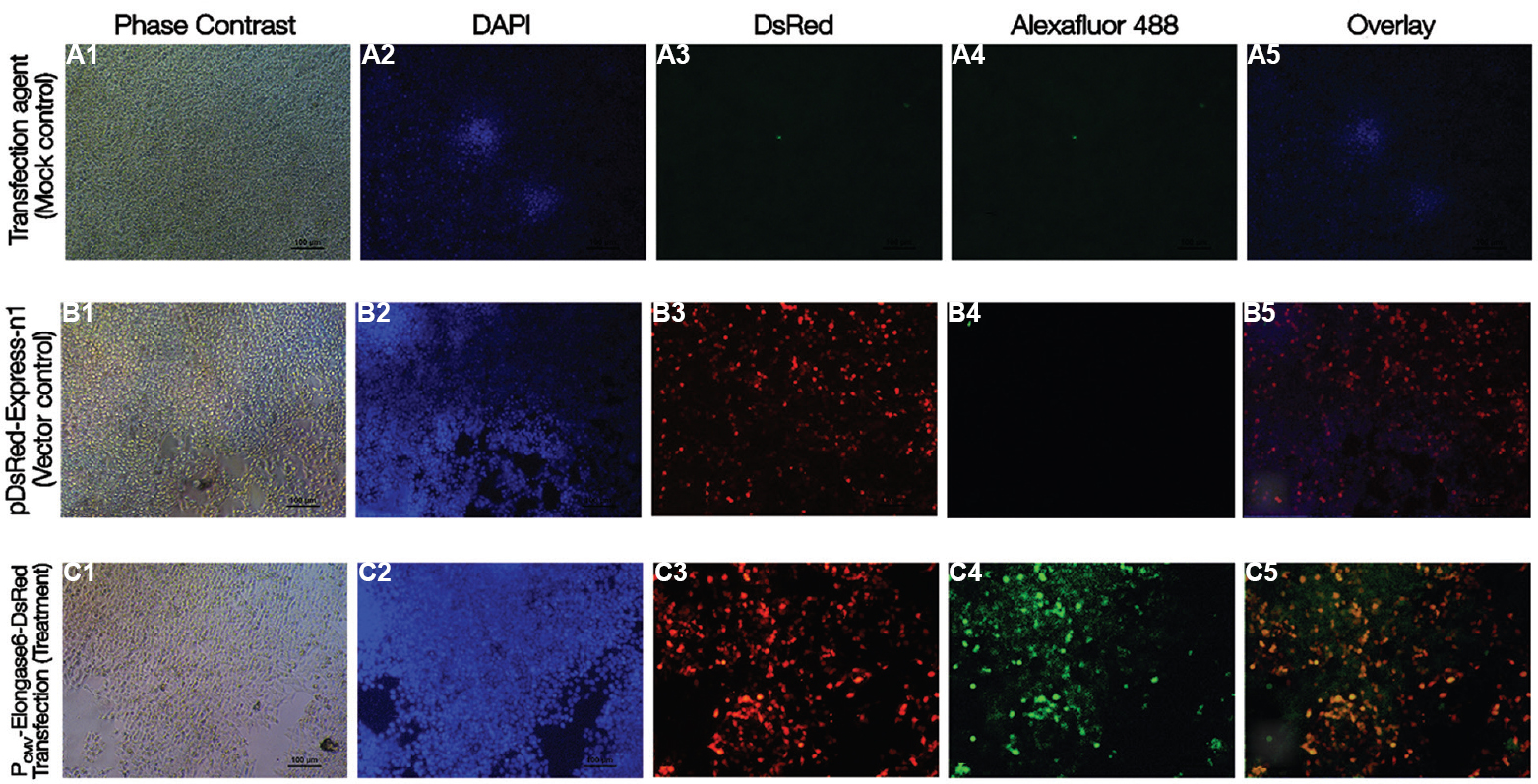
- Transient expression of recombinant porcine Elongase6 in HEK293T cells 48h post-transfection. HEK293T cells were grown and transfected with pDsRed-Express-N1 (empty vector), PCMV-Elongase6-DsRed (transgene expression vector) or sole polyethylenimine reagent (no plasmid, mock control). 48 h post-transfection, cells were fixed, immunostained to detect expression of Elongase6 protein (green) (for detail procedure, see text). Under a fluorescence microscope, cells expressing DsRed protein fluoresced red and nuclei stained with DAPI fluoresced blue. Photomicrograph of mock transfected HEK293T cells under bright field (A1), DAPI (blue) (A2), red channel (A3), green channel (A4), and overlay (A5) showed no fluorescence. For empty vector group, cells fluoresced blue (B2) and red (B3), not green (B4). (B1) The bright field image and overlay was shown in B5. However, in transgene expression vector group, cells exhibited blue (C2), red (C3) and green (C4) fluorescence. Phase contrast (C1) image and overlay of all different fluorescent channels (C5) are also shown. Scale bar=100μm.
Increasing evidence indicated that Elongase6 had a link to fatty acid metabolism and metabolic syndromes11161718. Downregulation of Elongase6 improved insulin resistance, independent of obesity in double knockout animal model10. However, Elongase6 overexpression in animal or cellular model has not been reported. This study described the generation of a transgene cassette for overexpression of porcine Elongase6 and demonstrated that the cassette was functional in human cell system. Thus, data presented here were a step forwards towards the generation of disease model overexpressing mammalian Elongase6.
It was further shown that Elongase6 of both human and porcine species shared a high degree (94% or more) of sequence identity at DNA and protein level, indicating higher degree of relatedness and shared homology in evolution. Absence of any amino acid residue likely to undergo glycosylation and no predicted B cell epitope in porcine Elongase6 indicated that expression of porcine Elongase6 might not elicit immunogenic response in host. Taken together, it seems that both human and porcine species possess structurally and functionally similar Elongase6.
Elongase6 was described as a rate-limiting enzyme regulating a number of downstream reactions. Given that, it would be essential to understand the entire regulatory network of Elongase6 for further identification of other potential candidates linked to metabolic disorder. In a separate experiment with adipogenic differentiation of MSCs, we demonstrated that ELOVL6 overexpression interfered with the master regulator, peroxisome proliferator-activated receptor gamma (data not shown). Therefore, the transgene construct presented here may serve as a seminal contribution towards constructing a regulatory network using an overexpression model and finding therapeutic targets for metabolic diseases. In addition, the biological activity of porcine ELOVL6 transgene could be assessed using the same differentiation experiment. Data from this experiment revealed that Elongase6 overexpression grossly interfered with differentiation process, affecting synthesis and organization of lipid droplets in adipose tissues (data not shown).
This fabricated gene construct would allow tracking intracellular trafficking and compartmental localization of Elongase6 by means of reporter fluorescent protein in suitable mammalian cells as well as in host tissues after transplantation. We chose MSCs, precursor stem cells for fat tissue1920 to clone and to express Elongase6 gene, following a two-step cloning strategy (Fig. 1B). The strategy might allow use each construct independently for other applications. CMV-IE promoter was used for strong and ubiquitous expression of the transgene. These strategies have been reported previously21.
Cloning of amplified product allowed expression of both Elongase6 and DsRed as a fusion protein, indicated by respective green and red fluorescence. Therefore, the gene construct was functional in HEK293T human cell line22, and it confirmed the activity of the porcine transgene in a heterologous system. Expressing any gene in a heterologous system sometimes poses an impediment that leads to no or altered product2324. In our study the codons of porcine Elongase6 were well recognized by the HEK293T cells. Perhaps the same construct would equally be effective for other mammalian cell system as well, owing to the presence of strong CMV-IE promoter25. This might help assess the effect of Elongase6 overexpression in a wide range of cell types.
Molecular cloning invariably involves the transformation of bacteria with plasmid containing cloned gene for the fact that bacteria are the most reliable host for maintaining and amplifying any gene carried by them. The expression cassette PCMV-Elongase6-DsRed was available as transformed DH5α bacteria strain, and this could be retrieved any time when needed. Although reporter protein DsRed is known to be functionally inert and does not interfere with cellular metabolism26, we suspect if the DsRed in fusion form with Elongase6 would gain any unprecedented activity. It is not clear if fusion with DsRed would alter (abate or potentiate) its functional activity as compared to the native Elongase6. Further, physical properties of the transgene derived Elongase6 should also be assessed. These issues need to be resolved in future experiments.
In conclusion, the present study described cloning and expression of porcine ELOVL6 gene in HEK293T cells. The data demonstrated that human cell, a heterologous system, supported the expression of porcine Elongase6. The fabricated gene cassette developed in this study might serve as an invaluable resource for deciphering underlying network associated with functions of Elongase6 and also for monitoring and/or modulating number of biological processes such as lipid metabolism, cellular homeostasis, differentiation of MSCs towards adipogenic lineage, development of metabolic disorders including insulin resistance, obesity and hepatosteatosis. This gene construct, if used in any forced-expression study, would be helpful to identify the set of upregulated genes in both human and swine species. These upregulated genes may help to frame a functional network within and between species and narrow down the field of investigation for further assessment of Elongase6 or other associated member of the network.
Acknowledgment
The work was supported by National Agricultural Science Fund, Indian Council of Agricultural Research (ICAR), New Delhi, India. Authors thank the Director, ICAR-Indian Veterinary Research Institute, Izatnagar, Bareilly, Uttar Pradesh, India, for his support.
Conflicts of Interest: None.
References
- Diabetes and cardiovascular disease in older adults: Current status and future directions. Diabetes. 2014;63:2578-89.
- [Google Scholar]
- Diabetes and hypertension: Is there a common metabolic pathway? Curr Atheroscler Rep. 2012;14:160-6.
- [Google Scholar]
- Elovl6 : A new player in fatty acid metabolism and insulin sensitivity. J Mol Med (Berl). 2009;87:379-84.
- [Google Scholar]
- “New” hepatic fat activates PPARalpha to maintain glucose, lipid, and cholesterol homeostasis. Cell Metab. 2005;1:309-22.
- [Google Scholar]
- Liver-specific deletion of acetyl-CoA carboxylase 1 reduces hepatic triglyceride accumulation without affecting glucose homeostasis. Proc Natl Acad Sci U S A. 2006;103:8552-7.
- [Google Scholar]
- Hepatic stearoyl-CoA desaturase-1 deficiency protects mice from carbohydrate-induced adiposity and hepatic steatosis. Cell Metab. 2007;6:484-96.
- [Google Scholar]
- Loss of stearoyl-CoA desaturase-1 function protects mice against adiposity. Proc Natl Acad Sci U S A. 2002;99:11482-6.
- [Google Scholar]
- Brown and beige fat : Development, function and therapeutic potential. Nat Med. 2013;19:1252-63.
- [Google Scholar]
- Role of fatty acid elongases in determination of de novo synthesized monounsaturated fatty acid species. J Lipid Res. 2010;51:1871-7.
- [Google Scholar]
- Crucial role of a long-chain fatty acid elongase, Elovl6, in obesity-induced insulin resistance. Nat Med. 2007;13:1193-202.
- [Google Scholar]
- Novel qualitative aspects of tissue fatty acids related to metabolic regulation : Lessons from Elovl6 knockout. Prog Lipid Res. 2012;51:267-71.
- [Google Scholar]
- Up-regulation of stearoyl-CoA desaturase 1 and elongase 6 genes expression in rat lipogenic tissues by chronic food restriction and chronic food restriction/refeeding. Mol Cell Biochem. 2010;345:181-8.
- [Google Scholar]
- Swine as models in biomedical research and toxicology testing. Vet Pathol. 2012;49:344-56.
- [Google Scholar]
- The production of multi-transgenic pigs: Update and perspectives for xenotransplantation. Transgenic Res. 2016;25:361-74.
- [Google Scholar]
- A comparative analysis of invasive and non-invasive method of bone marrow stromal cell isolation. Asian J Anim Vet Adv. 2015;10:549-55.
- [Google Scholar]
- The effect of LXRa, ChREBP and Elovl6 in liver and white adipose tissue on medium- and long-chain fatty acid diet-induced insulin resistance. Diabetes Res Clin Pract. 2013;102:183-92.
- [Google Scholar]
- Two modes of regulation of the fatty acid elongase ELOVL6 by the 3-ketoacyl-CoA reductase KAR in the fatty acid elongation cycle. PLoS One. 2014;9:e101823.
- [Google Scholar]
- Deletion of ELOVL6 blocks the synthesis of oleic acid but does not prevent the development of fatty liver or insulin resistance. J Lipid Res. 2014;55:2597-605.
- [Google Scholar]
- Human mesenchymal stem cells derived from limb bud can differentiate into all three embryonic germ layers lineages. Cell Reprogram. 2012;14:324-33.
- [Google Scholar]
- Mesenchymal stem cells: From biology to clinical use. Blood Transfus. 2007;5:120-9.
- [Google Scholar]
- Gene cloning and DNA analysis: An introduction (6th ed). West Sussex, UK: Wiley-Blackwell; 2015.
- Genome dynamics of the human embryonic kidney 293 lineage in response to cell biology manipulations. Nat Commun. 2014;5:4767.
- [Google Scholar]
- Influence of codon bias on heterologous production of human papillomavirus type 16 major structural protein L1 in yeast. ScientificWorldJournal 2012 2012:979218.
- [Google Scholar]
- Tissue and cell-specific transcriptional activity of the human cytomegalovirus immediate early gene promoter (UL123) in zebrafish. Gene Expr Patterns. 2013;13:91-103.
- [Google Scholar]
- Rapidly maturing variants of the discosoma red fluorescent protein (DsRed) Nat Biotechnol. 2002;20:83-7.
- [Google Scholar]





