Translate this page into:
Haplotype analysis of ADAM33 polymorphisms in asthma: A pilot study
For correspondence: Dr Padukudru A. Mahesh, Department of Respiratory Medicine, JSS Medical College, JSS Academy of Higher Education and Research (JSSAHER), Mysuru 570 015, Karnataka, India e-mail: mahesh1971in@yahoo.com
-
Received: ,
This is an open access journal, and articles are distributed under the terms of the Creative Commons Attribution-NonCommercial-ShareAlike 4.0 License, which allows others to remix, tweak, and build upon the work non-commercially, as long as appropriate credit is given and the new creations are licensed under the identical terms.
This article was originally published by Wolters Kluwer - Medknow and was migrated to Scientific Scholar after the change of Publisher.
Abstract
Background & objectives:
ADAM33 is implicated as a potentially strong candidate gene for asthma and bronchial hyper-responsiveness. Many polymorphisms of ADAM33 have been studied along with ADAM33 expression in various cells of the lungs. Haplotype analysis also showed association with asthma in different populations across the world. Therefore, the aim of this study was to perform a comprehensive screening of ADAM33 polymorphisms in adult patients with asthma.
Methods:
Thirty five polymorphisms of ADAM33 were genotyped in 55 patients with asthma and 53 controls. The association of single nucleotide polymorphisms (SNPs) and haplotypes with phenotypes of asthma was analysed.
Results:
The genotype, minor allele frequency, odds ratio and Hardy–Weinberg equilibrium did not show any significant difference among cases and controls. No association was found between SNPs of ADAM33 with the severity of asthma. Correlation analysis of ADAM33 SNPs to the phenotypes, based on clinical variables and allergen sensitization, did not show significant difference. Haplotype analysis showed that rs2280090 and rs2280091 were associated with asthma in the patient group.
Interpretation & conclusions:
Haplotype analysis showed an association of the two SNP variations with asthma. These SNPs lead to amino acid change and are prone to phosphorylation, which may affect expression levels and protein function of ADAM33 and asthma susceptibility.
Keywords
ADAM33
asthma
haplotype
polymorphisms
rs2280090
rs2280091
SNP
Asthma is associated with multiple single nucleotide polymorphisms (SNPs) of several genes; among them, ADAM33 is considered as a potentially significant asthma candidate gene. ADAM33 is one of the few genes that have been consistently found to be associated with asthma and bronchial hyper-responsiveness, and the findings have been reproduced in more than six independent populations; Caucasian, African-American, Hispanic, Japan, China, Korean and Jordan123. ADAM33 has been shown to have a higher expression in epithelial cells and smooth muscle cells of airways among asthmatics compared to controls. It has been identified as a potential therapeutic target for asthma as it enables airway inflammatory and remodelling effects after allergen exposure45. Every individual's genotype entails multiple closely linked SNPs, which can be referred to as haplotypes. Haplotypes are analyzed mainly for two reasons; primarily, haplotype alleles may exist in closer linkage disequilibrium with causal variant compared to single measured SNP and the other reason being that the haplotypes can be causal variants of significance by themselves6. The haplotype analysis can be considered to narrow down the position of disease susceptible loci, complex disease gene mapping and genome-wide association studies and to reconstruct the histories of human population78. Common haplotype frequencies can be evaluated based on the known marker phenotypes in unrelated individuals of a population9.
ADAM33 is important in asthma and is one of the few associations that have been successfully reproduced in follow up studies, including the haplotype analysis in different populations across the world. Over 55 different SNPs of ADAM33 have been reported in the literature, and the majority of these have been associated with asthma10. The present pilot study was carried out to comprehensively screen 35 ADAM33 polymorphisms that have been known to be associated with asthma in patients with asthma and healthy individuals.
Material & Methods
Consecutive patients with asthma (n=55) visiting the JSS Hospital, Mysuru, India, from July 2012 to January 2013 were included in the study. The age- and gender-matched controls were selected from the general population from the Burden of Obstructive Lung Disease (BOLD) cohort in urban Mysuru11. Both patients and normal controls underwent spirometry, pre- and post-bronchodilator, satisfying the American Thoracic Society standards12. The diagnosis of asthma was based according to the Global Initiative for Asthma (GINA) guidelines having a reversibility of more than 12 per cent and 200 ml on post-bronchodilator spirometry13. Asthma patients were classified according to severity based on the GINA-2012 guidelines as mild-, moderate- and severe-persistent asthma (https://ginasthma.org/wp-content/uploads/2019/01/2012-GINA.pdf). All the asthmatics underwent skin prick testing to confirm atopy. The patient was said to be atopic if he/she was sensitized to any one of the allergens tested. A wheal size of more than 3 mm of saline control was considered a positive skin prick test. Three millilitres of venous blood was collected into BD Vacutainer® Plus Plastic K2 EDTA tubes (USA). Wizard Genomic DNA isolation kit (Promega, USA) was used to isolate genomic DNA from peripheral blood leucocytes.
The patients below 18 yr were excluded from the study. Further, the patients with any other respiratory diseases were also excluded. The Institutional Human Ethical Committee of University of Mysore approved the present study (IHEC-UOM No.79 Ph.D/2012-13). Written informed consent was taken from all the patients/participants of the study.
Single nucleotide polymorphism (SNP) genotyping using MassARRAY analysis: SNP genotyping in the samples was performed using the Sequenom MassARRAY platform using MALDI-TOF mass spectrometry in Xcelris Genomics Company, Ahmedabad, India, wherein Sequenom-iPLEXR Gold SNP genotyping-platform (Agena Bioscience™, CA, USA) along with Spectro CHIP was used for the MassARRAY method and the analysis was done with MALDI-TOF mass spectrometry.
Analysis of SNPs of ADAM33 gene: ADAM33 consists of 22 exons and 21 introns encoding 813 amino acids. There are more than 340 SNPs identified in this gene14. This study was performed on 35 SNPs of ADAM33 selected based on previous studies1516 as well as from the databases (https://www.ncbi.nlm.nih.gov/snp/ and https://www.snpedia.com/index.php/SNPedia). The SNPs selected and their positions on the genome along with the type of SNPs, primers used for ADAM33 SNP genotyping alongside the mass of unextended (E1) and extended (E2) primers in Daltons are listed in Table I.
| SNP rs ID | Position in bp | Type of variant | UEP sequence | Allele (E1/E2) | Mass of UEP/E1/E2 (Dalton) |
|---|---|---|---|---|---|
| rs2280091 | 3669587 | Non-synonymous | GGGCGGCGTTCACCCCA | G/A | 5172.4/5419.5/5499.5 |
| rs2787094 | 3668514 | 3’UTR | GTCCACACTCCCCTG | G/C | 4448.9/4696.1/4736.1 |
| rs3918391 | 3675071 | Non-synonymous | CCAGGATACATAGAAACCCAC | C/T | 6377.2/6624.4/6704.3 |
| rs3918396 | 3671118 | Non-synonymous | GGCCATGCTCCTCAGC | A/G | 5107.3/5378.5/5394.5 |
| rs677044 | 3668784 | 3’UTR | CCAGCCCTCAGGAACTTCTA | G/A | 6021.9/6293.1/6309.1 |
| rs2853209 | 3670825 | Intronic | TGGCCTCCCAGTCAAGCG | T/A | 5764.7/6036/6091.8 |
| rs3918392 | 3674572 | Non-synonymous | GCTCACCTGGAAAGGA | A/G | 6176/6447.2/6463.2 |
| rs137919189 | 3668971 | Non-synonymous | GCTACCTCTCACCAGA | A/G | 5394.5/5665.7/5681.7 |
| rs41462450 | 3671022 | Non-synonymous | TGGGGCTGCAGAAGG | G/T | 5340.5/5627.7/5667.6 |
| rs6084434 | 3674821 | Synonymous | ACTACCAAGGGCGAGTAA | T/C | 5541.6/5812.8/5828.8 |
| rs3918394 | 3672848 | Non-synonymous | CCCCCCTTGCGGAAGAAG | T/C | 6970.5/7241.7/7257.7 |
| rs3918401 | 3668993 | Non-synonymous | TCCTCATTCTCAGCAGATCA | A/T | 6341.1/6612.3/6668.2 |
| rs2787093 | 3667815 | Downstream | GCAGGCCGAGCCTAG | T/C | 5470.6/5741.8/5757.8 |
| rs511898 | 3674438 | Intronic | TCGAGGCCTGTGAATTCC | C/T | 6775.4/7022.6/7102.5 |
| rs2485700 | 3674346 | Intronic | CTTCTGGGAGCTGGG | C/T | 6140/6411.2/6427.2 |
| rs2271510 | 3673503 | Intronic | CGAGTGGTCCTGGGG | A/G | 5595.6/5866.8/5882.8 |
| rs2485699 | 3674341 | Intronic | TGGGAGCTGGGATTGG | C/A | 5033.3/5320.5/5360.4 |
| rs574174 | 3670047 | Intronic | AATGACAAGGCCTTGGG | T/C | 5259.4/5530.6/5546.6 |
| rs12479696 | 3669310 | Non-synonymous | TGCCAGCAGTCTCGC | G/A | 5998.9/6246.1/6326 |
| rs615436 | 3672188 | Non-synonymous | GGGGCAGTGGCTACT | G/A | 4649/4896.2/4976.1 |
| rs17513846 | 3669893 | Intronic | TCCTCCAGGCTCTGA | C/A | 4503.9/4751.1/4775.1 |
| rs614971 | 3668203 | 3’UTR | ATCACCAGAGGCCAG | C/G | 4571/4818.2/4858.2 |
| rs41419248 | 3673394 | Synonymous | GGCTGCTGCGTGGAGGCT | G/T | 6214/6501.2/6541.1 |
| rs41534847 | 3673835 | Synonymous | GGACCGCAGCCGCGTCA | T/C | 5181.4/5428.6/5508.5 |
| rs41467948 | 3675034 | Non-synonymous | TGGTGCTGGCCCCCA | G/A | 5451.5/5722.7/5738.7 |
| rs41382144 | 3672615 | Non-synonymous | GTGAGCAAAGCAGCAGAG | G/C | 5935.9/6183.1/6223.1 |
| rs528557 | 3671095 | Synonymous | TGCCTCTGCTCCCAGG | G/C | 4809.1/5056.3/5096.3 |
| rs17548872 | 3669884 | Intronic | CTCTCTAGTCCTAACATTTCCTC | C/G | 6860.5/7107.7/7147.7 |
| rs41492952 | 3670152 | Intronic | GGCGTGTCAGCACGGA | C/G | 6577.3/6824.4/6864.5 |
| rs2280090 | 3669558 | Non-synonymous | CACAGCCACTGGACAG | G/A | 5486.6/5733.8/5813.7 |
| rs3918400 | 3668816 | 3’UTR | CTTCCCCGAGTGGAGCTT | T/C | 6414.2/6661.3/6741.3 |
| rs6084435 | 3682089 | 5’UTR | ACCCGTGCCCGGTGC | A/G | 5701.7/5972.9/5988.9 |
| rs11905233 | 3668529 | 3’UTR | CCCTATGGTTCGACTGA | T/C | 5161.4/5432.6/5448.6 |
| rs3746631 | 3668493 | 3’UTR | GGCTGGCCTCTGCAA | T/C | 4569/4840.2/4856.2 |
| rs517155 | 3668230 | 3’UTR | TGATCCTCCTACCCC | G/A | 4423.9/4695.1/4711.1 |
The “PED” format file with case-control values paired to the locus information file was considered for the haplotype analysis using Haploview tool (https://www.broadinstitute.org/haploview/haploview) which identified the association with T-int value. The association test was performed using the case/control possibility. The pairwise comparison of markers >500 kb distance was ignored followed by excluding individuals with >50 per cent missing genotypes. There were no individuals with missing genotype in our study dataset. The haplotype blocks were created with >1.0 per cent linked haplotype, and >10.0 per cent haplotype blocks were highlighted with the thick lines for association identification.
Effect of SNP variation: The possible effect of an amino acid change on the structure and function of a human protein can be predicted by the tool, PolyPhen-2 (Polymorphism Phenotyping V2; http://genetics.bwh.harvard.edu/pph2/). PolyPhen-2 uses eight sequence-based and three structure-based predictive features, which are selected automatically by an iterative greedy algorithm. The majority of these features involve comparison of a property of the wild-type (normal) allele and the corresponding property of the mutant (derived, disease-causing) allele, which together define an amino acid replacement17.
Statistical analysis: SNPs of ADAM33 were related with asthma using Hardy–Weinberg equilibrium, Chi-square test, odds ratio, ANOVA and Student's t test to understand the association of ADAM33 variations with asthma. These association tests between clinical variables and SNPs were tested using SPSS software V.19 (SPSS, Inc., Chicago, USA). Haploview software V.4.0 (Broad Institute, Cambridge, MA, USA) was used to assess the SNP haplotype and disease association.
Results
The demographic characteristics of the population studied are presented in Table II. Genotype to phenotype, minor allele frequency, odds ratio and Hardy–Weinberg equilibrium calculated did not show significant difference among cases and controls (data not shown). Thirty five polymorphisms of ADAM33 and their haplotypes were analysed for their association with asthma in 19 mild, 21 moderate and 15 severe cases. Table III represents the pulmonary function test values for both patients and controls. There was no association with any of the SNPs in mild, moderate or severe asthma as compared to controls. A comparison of the genotype distributions of ADAM33 SNPs among atopic and non-atopic patients showed an insignificant difference. Correlation analysis of ADAM33 SNPs using Chi square statistics to the clinical variables did not show positive correlation with the allergen sensitization, either with the number of allergen sensitization or with the severity of sensitization.
| Variables | Controls n=53 | Patients n=55 |
|---|---|---|
| Age (yr) | ||
| ≤40 | 35 (66.03) | 31 (56.36) |
| >40 | 18 (33.96) | 24 (43.63) |
| Gender | ||
| Male | 22 (41.50) | 20 (36.36) |
| Female | 31 (58.49) | 35 (63.63) |
| Family history of asthma | ||
| Yes | 0 | 24 (43.63) |
| No | 53 (100) | 31 (56.36) |
| Asthma duration (yr) | ||
| <1 | - | 4 (7.27) |
| 1-5 | - | 20 (36.36) |
| >5 | - | 31 (56.36) |
| Severity | ||
| Mild persistent | - | 19 (34.54) |
| Moderate persistent | - | 21 (38.18) |
| Severe persistent | - | 15 (27.27) |
| Atopic patients | - | 52 (94.54) |
| Non-atopic patients | - | 3 (5.45) |
Values in parenthesis are percentages
| Pulmonary function test parameters | Controls (pre-bronchodilator) (n=53) | Patients (pre-bronchodilator) (n=55) | Patients (post-bronchodilator) (n=55) |
|---|---|---|---|
| FVC (%) | 88.44±0.511 | 70.63±0.82 | 78.3±0.88 |
| FEV1 (%) | 88.84±0.47 | 69.12±0.89 | 78.81±0.72 |
| FEV1/FVC (%) | 104.23±0.35 | 99.54±0.62 | 106.83±0.53 |
| PEF (l/sec) | 92.81±0.95 | 74±1.0 | 84.56±0.90 |
Values are mean±SE. FEV1, forced expiratory volume in one second; FVC, forced vital capacity; PEF, peak expiratory flow
Haplotype analysis: Haplotype analysis showed that SNPs rs2280090 and rs2280091 were associated with asthma in the patient group (Table IV). The association of haplotype with asthma was analyzed using Haploview software similar to other studies181920. The haplotype with AA alleles was found to be significant (P=0.037) (Table V). The SNP location was 29 bp apart from each other and the LOD score was 20.48 followed by D --- 1. The r2 value was 0.75 and the distance between these SNPs was 29 bp. The Haploview software uses its own statistical value called T-int, which was found to be 105.54 (Table V and Fig. 1).
| L1 | L2 | D’ | LOD | r2 | CI-low | CI-hi | Dist | T-int |
|---|---|---|---|---|---|---|---|---|
| rs2787093 | rs3746631 | 0.016 | 0 | 0 | −0.01 | 0.28 | 678 | 6.91 |
| rs2787093 | rs2787094 | 1 | 5.28 | 0.213 | 0.7 | 1 | 699 | - |
| rs2787093 | rs677044 | 0.955 | 0.28 | 0.02 | 0.06 | 0.97 | 969 | - |
| rs2787093 | rs3918400 | 1 | 0.59 | 0.007 | 0.09 | 0.99 | 1001 | - |
| rs2787093 | rs2280090 | 1 | 0.76 | 0.017 | 0.12 | 0.99 | 1743 | - |
| rs2787093 | rs2280091 | 1 | 0.44 | 0.012 | 0.07 | 0.98 | 1772 | - |
| rs2787093 | rs17548872 | 1 | 0.59 | 0.007 | 0.09 | 0.99 | 2069 | - |
| rs2787093 | rs574174 | 1 | 1.14 | 0.022 | 0.18 | 0.99 | 2232 | - |
| rs2787093 | rs2853209 | 0.646 | 1 | 0.052 | 0.15 | 0.87 | 3010 | - |
| rs2787093 | rs528557 | 0.733 | 0.94 | 0.025 | 0.16 | 0.92 | 3280 | - |
| rs2787093 | rs3918396 | 1 | 0.4 | 0.012 | 0.07 | 0.98 | 3303 | - |
| rs2787093 | rs511898 | 0.776 | 1.26 | 0.029 | 0.22 | 0.94 | 6623 | - |
| rs2787093 | rs3918392 | 0.5 | 0.03 | 0.002 | 0.04 | 0.96 | 6757 | - |
| rs3746631 | rs2787094 | 0.711 | 2.67 | 0.13 | 0.37 | 0.88 | 21 | 21.21 |
| rs3746631 | rs677044 | 0.179 | 0.25 | 0.011 | 0.02 | 0.46 | 291 | - |
| rs3746631 | rs3918400 | 0.734 | 8.66 | 0.478 | 0.53 | 0.86 | 323 | - |
| rs3746631 | rs2280090 | 1 | 1.33 | 0.02 | 0.22 | 1 | 1065 | - |
| rs3746631 | rs2280091 | 1 | 0.95 | 0.015 | 0.14 | 0.99 | 1094 | - |
| rs3746631 | rs17548872 | 0.734 | 8.66 | 0.478 | 0.53 | 0.86 | 1391 | - |
| rs3746631 | rs574174 | 0.68 | 3.49 | 0.165 | 0.39 | 0.85 | 1554 | - |
| rs3746631 | rs2853209 | 0.79 | 1.44 | 0.038 | 0.25 | 0.94 | 2332 | - |
| rs3746631 | rs528557 | 0.608 | 1.26 | 0.061 | 0.18 | 0.83 | 2602 | - |
| rs3746631 | rs3918396 | 1 | 0.3 | 0.014 | 0.06 | 0.98 | 2625 | - |
| rs3746631 | rs511898 | 0.337 | 0.38 | 0.018 | 0.04 | 0.66 | 5945 | - |
| rs3746631 | rs3918392 | 0.316 | 1.69 | 0.088 | 0.12 | 0.52 | 6079 | - |
| rs2787094 | rs677044 | 0.039 | 0.03 | 0.001 | −0.01 | 0.25 | 270 | 34.15 |
| rs2787094 | rs3918400 | 0.784 | 3.01 | 0.14 | 0.43 | 0.92 | 302 | - |
| rs2787094 | rs2280090 | 1 | 3.02 | 0.078 | 0.53 | 1 | 1044 | - |
| rs2787094 | rs2280091 | 1 | 2.57 | 0.058 | 0.47 | 1 | 1073 | - |
| rs2787094 | rs17548872 | 0.784 | 3.01 | 0.14 | 0.43 | 0.92 | 1370 | - |
| rs2787094 | rs574174 | 0.765 | 10.76 | 0.421 | 0.61 | 0.87 | 1533 | - |
| rs2787094 | rs2853209 | 0.393 | 0.7 | 0.037 | 0.07 | 0.66 | 2311 | - |
| rs2787094 | rs528557 | 0.433 | 2.39 | 0.12 | 0.21 | 0.6 | 2581 | - |
| rs2787094 | rs3918396 | 0.762 | 5.69 | 0.23 | 0.52 | 0.89 | 2604 | - |
| rs2787094 | rs511898 | 0.283 | 0.95 | 0.049 | 0.07 | 0.48 | 5924 | - |
| rs2787094 | rs3918392 | 0.343 | 0.54 | 0.027 | 0.05 | 0.64 | 6058 | - |
| rs677044 | rs3918400 | 0.448 | 1.3 | 0.062 | 0.14 | 0.69 | 32 | 52.37 |
| rs677044 | rs2280090 | 1 | 0.77 | 0.057 | 0.12 | 0.99 | 774 | - |
| rs677044 | rs2280091 | 0.192 | 0.02 | 0.002 | 0.03 | 0.94 | 803 | - |
| rs677044 | rs17548872 | 0.448 | 1.3 | 0.062 | 0.14 | 0.69 | 1100 | - |
| rs677044 | rs574174 | 0.002 | 0 | 0 | 0 | 0.53 | 1263 | - |
| rs677044 | rs2853209 | 0.266 | 0.81 | 0.031 | 0.05 | 0.48 | 2041 | - |
| rs677044 | rs528557 | 0.094 | 0.04 | 0.001 | 0 | 0.46 | 2311 | - |
| rs677044 | rs3918396 | 0.677 | 0.66 | 0.019 | 0.11 | 0.91 | 2334 | - |
| rs677044 | rs511898 | 0.176 | 0.15 | 0.005 | 0.01 | 0.5 | 5654 | - |
| rs677044 | rs3918392 | 1 | 8.58 | 0.307 | 0.78 | 1 | 5788 | - |
| rs3918400 | rs2280090 | 1 | 0.85 | 0.018 | 0.13 | 0.99 | 742 | 75.37 |
| rs3918400 | rs2280091 | 1 | 0.51 | 0.013 | 0.08 | 0.98 | 771 | - |
| rs3918400 | rs17548872 | 1 | 20.74 | 1 | 0.9 | 1 | 1068 | - |
| rs3918400 | rs574174 | 1 | 8.18 | 0.316 | 0.77 | 1 | 1231 | - |
| rs3918400 | rs2853209 | 1 | 2.33 | 0.054 | 0.43 | 1 | 2009 | - |
| rs3918400 | rs528557 | 1 | 3.97 | 0.145 | 0.63 | 1 | 2279 | - |
| rs3918400 | rs3918396 | 1 | 0.47 | 0.013 | 0.08 | 0.98 | 2302 | - |
| rs3918400 | rs511898 | 0.849 | 2.31 | 0.101 | 0.4 | 0.96 | 5622 | - |
| rs3918400 | rs3918392 | 0.535 | 4.79 | 0.287 | 0.33 | 0.7 | 5756 | - |
| rs2280090 | rs2280091 | 1 | 20.48 | 0.75 | 0.89 | 1 | 29 | 105.54 |
| rs2280090 | rs17548872 | 1 | 0.85 | 0.018 | 0.13 | 0.99 | 326 | - |
| rs2280090 | rs574174 | 1 | 1.27 | 0.056 | 0.21 | 0.99 | 489 | - |
| rs2280090 | rs2853209 | 0.451 | 1.61 | 0.066 | 0.17 | 0.66 | 1267 | - |
| rs2280090 | rs528557 | 0.891 | 8.2 | 0.279 | 0.69 | 0.97 | 1537 | - |
| rs2280090 | rs3918396 | 0.564 | 0.13 | 0.01 | 0.05 | 0.96 | 1560 | - |
| rs2280090 | rs511898 | 0.777 | 5.85 | 0.204 | 0.54 | 0.9 | 4880 | - |
| rs2280090 | rs3918392 | 0.938 | 0.25 | 0.016 | 0.06 | 0.97 | 5014 | - |
| rs2280091 | rs17548872 | 1 | 0.51 | 0.013 | 0.08 | 0.98 | 297 | 83.22 |
| rs2280091 | rs574174 | 1 | 1.16 | 0.042 | 0.19 | 0.99 | 460 | - |
| rs2280091 | rs2853209 | 0.573 | 1.71 | 0.08 | 0.22 | 0.78 | 1238 | - |
| rs2280091 | rs528557 | 0.926 | 6.35 | 0.226 | 0.68 | 0.98 | 1508 | - |
| rs2280091 | rs3918396 | 1 | 0.38 | 0.023 | 0.07 | 0.98 | 1531 | - |
| rs2280091 | rs511898 | 0.774 | 4.1 | 0.152 | 0.48 | 0.91 | 4851 | - |
| rs2280091 | rs3918392 | 0.334 | 0.03 | 0.001 | 0.03 | 0.95 | 4985 | - |
| rs17548872 | rs574174 | 1 | 8.18 | 0.316 | 0.77 | 1 | 163 | 66.94 |
| rs17548872 | rs2853209 | 1 | 2.33 | 0.054 | 0.43 | 1 | 941 | - |
| rs17548872 | rs528557 | 1 | 3.97 | 0.145 | 0.63 | 1 | 1211 | - |
| rs17548872 | rs3918396 | 1 | 0.47 | 0.013 | 0.08 | 0.98 | 1234 | - |
| rs17548872 | rs511898 | 0.849 | 2.31 | 0.101 | 0.4 | 0.96 | 4554 | - |
| rs17548872 | rs3918392 | 0.535 | 4.79 | 0.287 | 0.33 | 0.7 | 4688 | - |
| rs574174 | rs2853209 | 0.803 | 3.06 | 0.111 | 0.45 | 0.93 | 778 | 99.98 |
| rs574174 | rs528557 | 0.958 | 12.96 | 0.422 | 0.81 | 0.99 | 1048 | - |
| rs574174 | rs3918396 | 1 | 15.58 | 0.55 | 0.87 | 1 | 1071 | - |
| rs574174 | rs511898 | 0.913 | 10.64 | 0.368 | 0.74 | 0.97 | 4391 | - |
| rs574174 | rs3918392 | 0.455 | 1.39 | 0.065 | 0.15 | 0.7 | 4525 | - |
| rs2853209 | rs528557 | 0.014 | 0 | 0 | −0.01 | 0.2 | 270 | 80.79 |
| rs2853209 | rs3918396 | 0.59 | 0.76 | 0.033 | 0.11 | 0.85 | 293 | - |
| rs2853209 | rs511898 | 0.004 | 0 | 0 | −0.01 | 0.18 | 3613 | - |
| rs2853209 | rs3918392 | 1 | 2.63 | 0.054 | 0.47 | 1 | 3747 | - |
| rs528557 | rs3918396 | 1 | 8.27 | 0.253 | 0.8 | 1 | 23 | 80.25 |
| rs528557 | rs511898 | 0.896 | 28.4 | 0.771 | 0.81 | 0.95 | 3343 | - |
| rs528557 | rs3918392 | 0.357 | 0.5 | 0.018 | 0.05 | 0.67 | 3477 | - |
| rs3918396 | rs511898 | 0.922 | 6.07 | 0.207 | 0.67 | 0.98 | 3320 | 56.9 |
| rs3918396 | rs3918392 | 0.865 | 0.17 | 0.01 | 0.05 | 0.97 | 3454 | - |
| rs511898 | rs3918392 | 0.338 | 0.43 | 0.016 | 0.04 | 0.66 | 134 | 5.12 |
D’, D prime between two loci; LOD, log of likelihood odds ratio; r2, correlation coefficient between two loci; CI-low and CI-hi, 95% confidence interval lower and upper bound on D; Dist, Distance between two loci; T-int, statistic used by HapMap project in Haploview software to measure the completeness of information represented by a set of markers in a region are represented
| SNPs | Block | χ2 | Case-control frequency | Haplotype (P) |
|---|---|---|---|---|
| rs2280090 rs2280091 |
GA | 2.256 | 0.791, 0.868 | 0.829 |
| AG | 0.794 | 0.155, 0.113 | 0.134 | |
| AA | 1.927 | 0.055, 0.019 | 0.037 | |
| D’ - 1, LOD - 20.48, r2-0.75, Distance - 29 bp, T-int - 105.54 | ||||
D’, D prime between two loci; LOD, log of likelihood odds ratio; r2, correlation coefficient between two loci
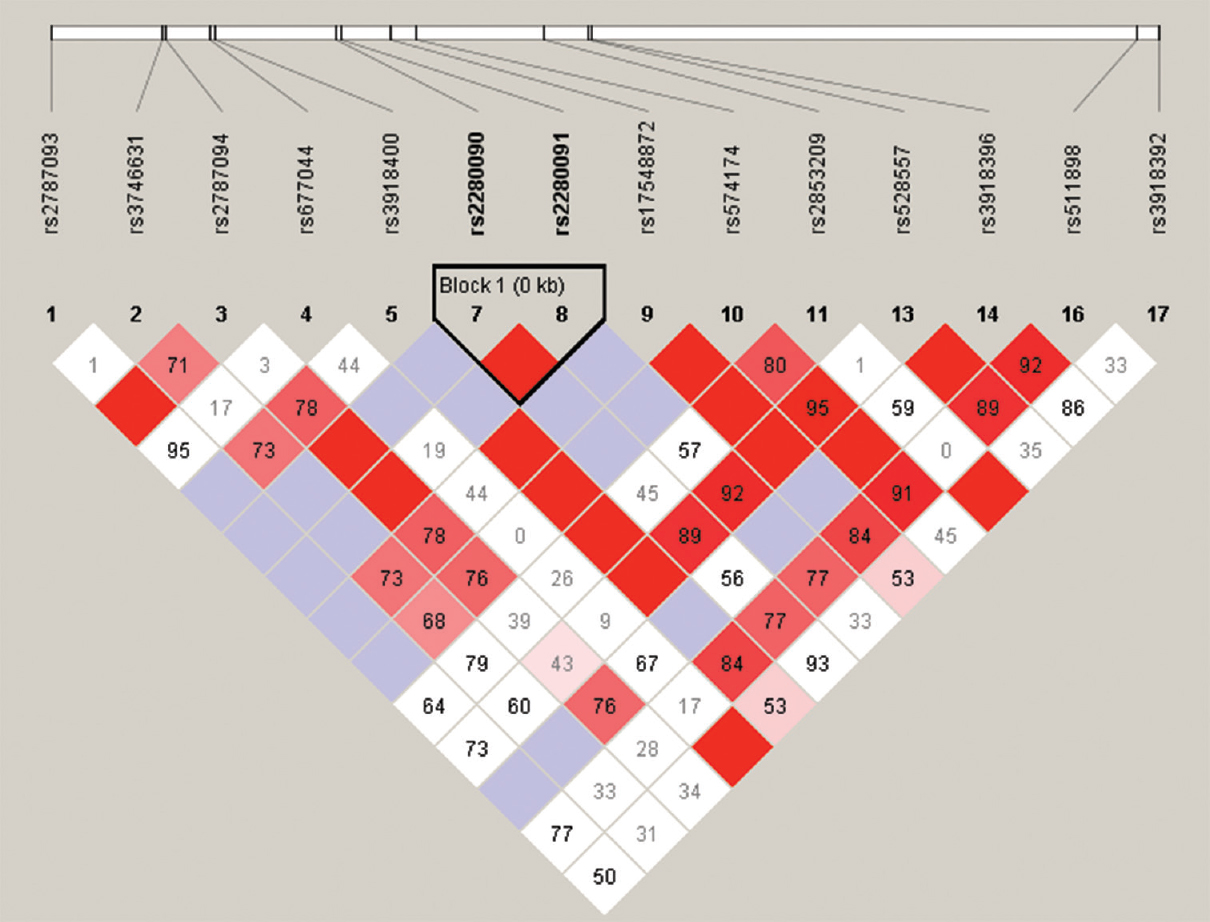
- Haplotype blocks showing association between asthma and ADAM33 polymorphisms rs2280090 and rs2280091. The two SNPs are 29 bp apart from each other with a LOD score of 20.48.
PolyPhen-2 analysis of significantly associated variations showed the effect 'benign' for both rs2280090 and rs2280091 SNPs individually (Fig. 2).

- PolyPhen-2 analysis showing the “Benign” effect prediction individually with (A) a score of 0.081 for the SNP rs2280091 (sensitivity: 0.93 and specificity: 0.85) and, (B) a score of 0.036 for the SNP rs2280090 (sensitivity: 0.94 and specificity: 0.82).
Discussion
Asthma is a heterogeneous, chronic lung disease that is caused by interplay of genes and environment in an individual. ADAM33 is one of the positionally cloned asthma-associated genes found to be associated with many pathological features of asthma, including goblet cell hyperplasia, subepithelial fibrosis, collagen deposition, mucosal gland hyperplasia, smooth muscle hypertrophy, changes in the extracellular matrix and inflammation42122. In the present study the genotype to phenotype, minor allele frequency, and odds ratios did not show significant difference among cases and controls with SNPs. On haplotype analysis, an association of two SNPs rs2280090 and rs2280091 was observed with asthma with a strong linkage disequilibrium value.
Many SNPs associated with asthma have not been found to be reproducible in different ethnic populations such as Puerto Rican and Mexican23. ADAM33 SNPs have been found to be associated with asthma susceptibility in various ethnic populations, though with different frequencies, and are one of the most reproducible variables with odds ratio 1.67 to 4.34 (e.g., US White and US Hispanic, Black Americans, UK, Europeans and Asians)24.
The study on ADAM33 polymorphisms was carried out as continuation of studies in genetic polymorphisms in asthma by the Mysore asthma genetics group22526. No association was found in our study between ADAM33 SNPs and asthma phenotypes such as asthma severity and atopy. Our results were similar to other studies conducted in Australian and German populations127. On the other hand, in a study from north India28 one of the SNPs of ADAM33 was found associated with the mild intermittent subgroup of asthma28.
On haplotype analysis, two polymorphisms, rs2280090 and rs2280091, were observed to be associated with asthma among 35 SNPs, with the T-int value of 105.54. However, Haploview software measures the completeness of information represented by a set of markers in a region and denoted as T-int. The value of T-int >100 is considered as significantly associated as per the Haploview tool. The result supported another study where haplotypes were found to be associated with asthma18; further, these SNPs were associated with an increased predisposition to asthma29, but no such association was found in other studies3031. In another study, rs2853209 SNP was found to be associated with asthma along with T1 (rs2280091) and F+1 SNPs of ADAM3332.
A similar study on Korean asthmatics confirmed that both SNPs and SNP haplotypes were associated with bronchial hyper-responsiveness and asthma33. No haplotype in linkage disequilibrium was consistent in all populations studied. This could be due to differences in each population's linkage disequilibrium34. The minor alleles observed in the two SNPs in the present study (rs2280090 and rs2280091) were in the linkage group with a strong association of 20.48 LOD score. These two SNPs formed the haplotype block and strong LOD value supported the association of this haplotype with asthma.
The two polymorphisms associated with asthma in our study were located on the exonic region of the ADAM33. The rs2280090 is a variation wherein the amino acid proline changes to serine and the other SNP rs2280091 naturally codes for amino acid methionine changing to threonine. Both the SNPs are located on the cytoplasmic domain of the gene, and these variations have a potential to change the intracellular signal of the protein that contributes to increasing fibroblast and proliferation of smooth muscles. These two features are characteristic of airway remodelling in asthma2934.
These variations were found to be benign in their effect individually, with respect to the predictions of PolyPhen-2 predictor tool. The effect of phosphorylation on the amino acids serine and threonine is high, and these amino acid residues mediate numerous signal transduction pathways3536. Since these variations were found to be together in the participants of the study, these might have combined effect on the protein function.
The study was performed as a pilot study with a limited sample number, which remained the main limitation of this study. The inclusion of 35 SNPs allowed us to perform the haplotype analysis. An association of the two SNPs on haplotype analysis was observed linked with each other with strong LOD score, and since both the SNPs are located on the same domain (cytoplasmic domain) of the ADAM33 protein, it is possible that it may hamper the protein function. This suggests that further functional analysis of this domain may help us to better understand the role of ADAM33 in asthma.
In conclusion, an association of two polymorphisms (rs2280090 and rs2280091) was found with asthma on haplotype analysis but not to other asthma phenotypes, such as asthma severity and atopy. Since this was a pilot study, the sample size used for the study was small and further studies with larger sample size would be necessary.
Acknowledgment
Authors acknowledge JSS Hospital and Medical College, University of Mysore, for providing facility to conduct this work and Asthma Allergy Associates, Mysuru and Genetics and Genomics Laboratory, University of Mysore, Mysuru, for support.
Financial support & sponsorship: Authors acknowledge the funding agency, Indian Council of Medical Research (RFC No: NCD-Ad-hoc/3/2011-12), and Council of Scientific and Industrial Research [No.60(0098)/11/EMR-II], New Delhi, for financial support.
Conflicts of Interest: None.
References
- ADAM33 haplotypes are associated with asthma in a large Australian population. Eur J Hum Genet. 2006;14:1027-36.
- [Google Scholar]
- Association of IL-4 and ADAM33 gene polymorphisms with asthma in an Indian population. Lung. 2010;188:415-22.
- [Google Scholar]
- Role of ADAM33 gene and associated single nucleotide polymorphisms in asthma. Allergy Rhinol (Providence). 2011;2:e63-70.
- [Google Scholar]
- Multi-SNP haplotype analysis methods for association analysis. Methods Mol Biol. 2012;850:423-52. 6
- [Google Scholar]
- The accuracy of statistical methods for estimation of haplotype frequencies: An example from the CD4 locus. Am J Hum Genet. 2000;67:518-22.
- [Google Scholar]
- Effectiveness of computational methods in haplotype prediction. Hum Genet. 2002;110:148-56.
- [Google Scholar]
- Efficiency of estimation of haplotype frequencies: Use of marker phenotypes of unrelated individuals versus counting of phase-known gametes. Am J Hum Genet. 2000;67:1626-7.
- [Google Scholar]
- An understanding of the genetic basis of asthma. Indian J Med Res. 2011;134:149-61.
- [Google Scholar]
- Allergy asthma practice in India: Beyond the guidelines “Shivpuri Oration 2017”. Indian J Allergy Asthma Immunol. 2019;33:8-13.
- [Google Scholar]
- American Thoracic society, guidelines. https://www.thoracic.org/professionals/clinical-resources/asthma-center/guidelines.php
- Pocket guide for asthma management and prevention (for adults and children older than 5 years). Fontana, WI: Global Initiative for Asthma; 2011. p. :1-32.
- Association between genetic polymorphisms in the ADAM33 gene and asthma risk: A meta-analysis. DNA Cell Biol. 2014;33:793-801.
- [Google Scholar]
- Association of IL-4 and ADAM33 gene polymorphisms with asthma in an Indian population. Lung. 2010;188:415-22.
- [Google Scholar]
- Role of ADAM33 gene and associated single nucleotide polymorphisms in asthma. Allergy Rhinol (Providence). 2011;2:e63-70.
- [Google Scholar]
- Predicting functional effect of human missense mutations using polyPhen-2. Curr Protoc Hum Genet. 2013;76:20-41.
- [Google Scholar]
- A six-SNP haplotype of ADAM33 is associated with asthma in a population of Cartagena, Colombia. Int Arch Allergy Immunol. 2010;152:32-40.
- [Google Scholar]
- Association of toll-like receptor 4 polymorphisms with diabetic foot ulcers and application of artificial neural network in DFU risk assessment in type 2 diabetes patients. Biomed Res Int. 2013;2013:318686.
- [Google Scholar]
- Human ADAM33 messenger RNA expression profile and post-transcriptional regulation. Am J Respir Cell Mol Biol. 2003;29:571-82.
- [Google Scholar]
- Molecular interaction network and pathway studies of ADAM33 potentially relevant to asthma. Ann Allergy Asthma Immunol. 2014;113:418-240.
- [Google Scholar]
- ADAM33 is not associated with asthma in Puerto Rican or Mexican populations. Am J Respir Crit Care Med. 2003;168:1312-6.
- [Google Scholar]
- ADAM metallopeptidase domain 33 (ADAM33): a promising target for asthma. Mediators Inflamm. 2014;2014:572025.
- [Google Scholar]
- Serum levels of interleukin-13 and interferon-gamma from adult patients with asthma in Mysore. Cytokine. 2012;60:431-7.
- [Google Scholar]
- Serum levels of IL-10, IL-17F and IL-33 in patients with asthma: A case-control study. J Asthma. 2014;51:1004-13.
- [Google Scholar]
- The role of polymorphisms in ADAM33, a disintegrin and metalloprotease 33, in childhood asthma and lung function in two German populations. Respir Res. 2006;7:91.
- [Google Scholar]
- Association of ADAM33 gene polymorphisms with adult-onset asthma and its severity in an Indian adult population. J Genet. 2011;90:265-73.
- [Google Scholar]
- T1 and T2 ADAM33 single nucleotide polymorphisms and the risk of childhood asthma in a Saudi Arabian population: A pilot study. Ann Saudi Med. 2012;32:479-86.
- [Google Scholar]
- Association of ADAM33 gene polymorphisms with allergic asthma. Iran J Basic Med Sci. 2014;17:716-21.
- [Google Scholar]
- Possible control of paternal imprinting of polymorphisms of the ADAM33 gene by epigenetic mechanisms and association with level of airway hyperresponsiveness in asthmatic children. Biomed Pap Med Fac Univ Palacky Olomouc Czech Repub. 2013;157:367-73.
- [Google Scholar]
- Association of ADAM33 gene polymorphisms with asthma in the Uygur population of China. Biomed Rep. 2013;1:447-53.
- [Google Scholar]
- ADAM33 polymorphism: Association with bronchial hyper-responsiveness in Korean asthmatics. Clin Exp Allergy. 2004;34:860-5.
- [Google Scholar]
- Association of a disintegrin and metalloprotease 33 (ADAM33) gene with asthma in ethnically diverse populations. J Allergy Clin Immunol. 2003;112:717-22.
- [Google Scholar]
- The regulation of protein function by multisite phosphorylation – A 25 year update. Trends Biochem Sci. 2000;25:596-601.
- [Google Scholar]
- Phosphorylation of basic amino acid residues in proteins: Important but easily missed. Acta Biochim Pol. 2011;58:137-48.
- [Google Scholar]






