Translate this page into:
Haitian variant tcpA in Vibrio cholerae O1 El Tor strains in National Capital Region (India)
*For correspondence: sharmanc1@rediffmail.com
-
Received: ,
This is an open access article distributed under the terms of the Creative Commons Attribution-NonCommercial-ShareAlike 3.0 License, which allows others to remix, tweak, and build upon the work non-commercially, as long as the author is credited and the new creations are licensed under the identical terms.
This article was originally published by Medknow Publications & Media Pvt Ltd and was migrated to Scientific Scholar after the change of Publisher.
Sir,
Vibrio cholerae O1 is the causative agent of cholera, which has two biotypes, namely, classical and El Tor, based on various phenotypic and genotypic characters1. Cholera toxin (ctx) and toxin co-regulated pilus (tcp) are essential virulence genes. The expression of CT and TCP is regulated by ToxR, a co-regulatory protein2. TCP is a type IV pilus which is essential for colonization in the small intestine3. Whole genome sequence of V. cholerae strains isolated from Bangladesh (CIRS101) and Haitian outbreak has shown a single nucleotide polymorphism (SNP) at a nucleotide position 266 (amino acid 89) of the tcpA gene, particularly associated with the Haitian variant456. We undertook this study to investigate the presence of mutation in tcpA allele in V. cholerae El Tor strains obtained from National Capital Region (NCR) of India.
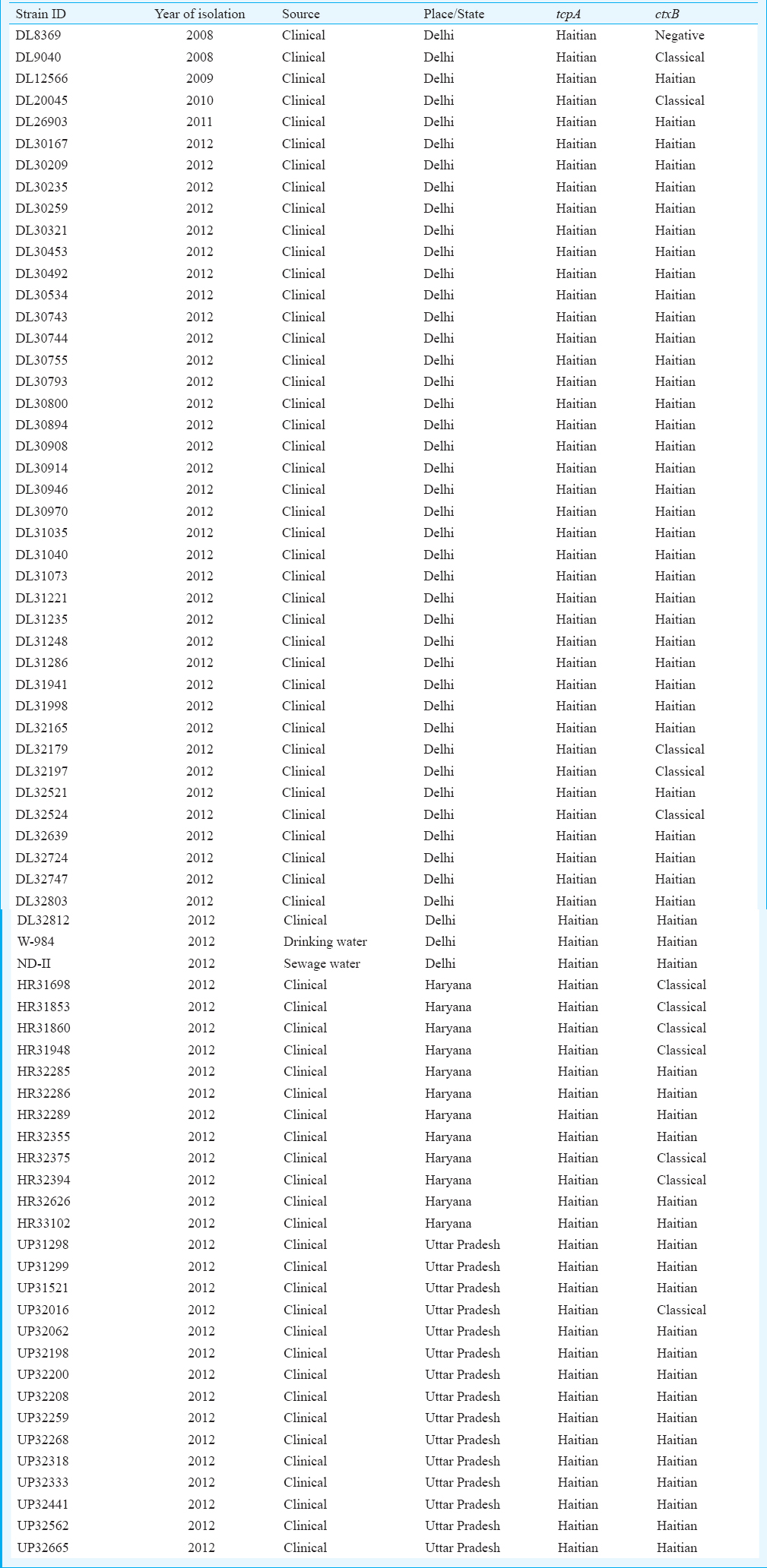
A total of 71 V. cholerae strains could be revived from the collection maintained in the Laboratory Department of Maharishi Valmiki Infectious Diseases Hospital (MVIDH), Delhi, India. These strains were collected from NCR (Delhi, Haryana and Uttar Pradesh) during 2008-2012 and were characterized biochemically and serotyped as V. cholerae O1 Ogawa using standard procedure7. Environmental sampling and processing of samples were done according to Mishra et al8. A total of 204 samples were collected which included 171 drinking water samples from the houses of patients admitted to MVIDH, 13 samples from Najafgarh drain, seven samples from different lakes and three samples from the Yamuna river. Only two drinking water samples and one sewage samples were positive for V. cholerae O1. Detection of the mutation in tcpA gene encoding TCP was done by a polymerase chain reaction (PCR) assay9. This PCR assay discriminates the V. cholerae strains harbouring Haitian, classical and El Tor alleles of tcpA, and this may be used to understand the presence of the new variant in different areas of cholera endemicity.
In this study, three different primers used included one common reverse primer for both El Tor and Haitian type tcpA alleles [tcpA El-Rev (5’-CCGACTGTAATTGCGAATGC-3’)]. Two forward primers [tcpA-F’1 (5’-CCAGCTACCGCAAACGCAGA-3’) and tcpA-F’2 (5’-CCAGCTACCGCAAACGCAGG-3’)] specific for El Tor and Haitian type tcpA alleles were used, respectively. The PCR assay conditions and PCR cycles were as described previously9. N16961 was used as a control strain for El Tor, and EL-1786 for Haitian, to check the mutation in tcpA gene. These control strains were obtained from National Institute of Cholera and Enteric Diseases (NICED), Kolkata, India.
PCR assay confirmed all the 71 strains carrying tcpA of Haitian type which yielded a 167bp fragment with Haitian-specific primer pair but not with El Tor-specific primer. Only V. cholerae O1 Inaba El Tor biotype (control strain N16961) was amplified with El Tor-specific primers which yielded a 167bp fragment but not with Haitian tcpA-specific primer. Previous studies reported a single nucleotide change at 266 position which resulted in asparagine to serine substitution49. This mutation (Asn→Ser) at the 89th amino acid of whole tcpA was the result of transition which took place in isolates of Bangladesh in 200210 and isolates of Kolkata, India, in 20039. Haitian tcpA allele (tcpETCIRS) has been found among the isolates of Afghanistan, Cameroon, India, Nepal, Nigeria, Pakistan, South Africa and Sri Lanka5. In this study, the combination of Haitian ctxB (ctxB7), classical ctxB (ctxB1) and tcpA of Haitian allele is reported for the first time since 2008 in north India including Delhi, Haryana and Uttar Pradesh in both 69 clinical and two environmental isolates (sewage & drinking water) (Table). The presence of tcpA Haitian allele was not restricted to strains having ctxB7 allele. It was equally present among both ctxB1 and ctxB7 isolates. Such molecular intricacies are important to understand the contemporary developments taking place in ctxB and tcpA genes globally. Keeping in view of the Haiti experiences and findings in our country, there is a need for the redressal of control strategies being adopted in the surveillance of cholera disease in this endemic region.
Acknowledgment
The first author (DK) was a recipient of RGNF fellowship from the University Grants Commission (UGC), New Delhi.
Conflicts of Interest: None.
References
- Development and validation of a mismatch amplification mutation PCR assay to monitor the dissemination of an emerging variant of Vibrio cholerae O1 biotype El Tor. Microbiol Immunol. 2008;52:314-7.
- [Google Scholar]
- Epidemiology, genetics, and ecology of toxigenic Vibrio cholerae. Microbiol Mol Biol Rev. 1998;62:1301-14.
- [Google Scholar]
- Toxin, toxin-coregulated pili, and the toxR regulon are essential for Vibrio cholerae pathogenesis in humans. J Exp Med. 1988;168:1487-92.
- [Google Scholar]
- Genome sequence of hybrid Vibrio cholerae O1 MJ-1236, B-33, and CIRS101 and comparative genomics with V. cholerae. J Bacteriol. 2010;192:3524-33.
- [Google Scholar]
- Characterization of toxigenic Vibrio cholerae from Haiti, 2010-2011. Emerg Infect Dis. 2011;17:2122-9.
- [Google Scholar]
- World Health Organization. Manual for the Laboratory Identification and Antimicrobial Susceptibility Testing of Bacterial Pathogens of Public Health Importance in the Developing World. Centers for Disease Control and Prevention, Atlanta Georgia USA. USAID/WHO/CDC 2003:141-59.
- Amplified fragment length polymorphism of clinical and environmental Vibrio cholerae from a freshwater environment in a cholera-endemic area, India. BMC Infect Dis. 2011;11:249.
- [Google Scholar]
- Haitian variant tcpA in Vibrio cholerae O1 El Tor strains in Kolkata, India. J Clin Microbiol. 2014;52:1020-1.
- [Google Scholar]
- Genomic diversity of 2010 Haitian cholera outbreak strains. Proc Natl Acad Sci USA. 2012;109:E2010-7.
- [Google Scholar]





