Translate this page into:
Genetic diversity study of various β-lactamase-producing multidrug-resistant Escherichia coli isolates from a tertiary care hospital using ERIC-PCR
Reprint requests: Dr. Enketeswara Subudhi, Centre of Biotechnology, Siksha ‘O’ Anusandhan University, Khandagiri, Bhubaneswar 751 003, Odisha, India e-mail: enketeswarasubudhi@soauniversity.ac.in
-
Received: ,
This is an open access article distributed under the terms of the Creative Commons Attribution-NonCommercial-ShareAlike 3.0 License, which allows others to remix, tweak, and build upon the work non-commercially, as long as the author is credited and the new creations are licensed under the identical terms.
This article was originally published by Medknow Publications & Media Pvt Ltd and was migrated to Scientific Scholar after the change of Publisher.
Abstract
Background & objectives:
The prevalence of multidrug-resistant (MDR) Escherichia coli isolates producing β-lactamase enzyme is a growing problem across the globe. Strain typing is an epidemiologically important tool not only for detecting the cross transmission of nosocomial pathogens but also for determining the source of infection. The present study was conducted to understand the clonal relationship among various β-lactamase-producing MDR E. coli isolates using enterobacterial repetitive intergenic consensus (ERIC) polymerase chain reaction (PCR).
Methods:
A total of 41 MDR E. coli isolates were randomly collected from various clinical samples and processed. Isolated organisms were tested for antibiotics resistance pattern. Phenotypic detection of metallo β-lactamases (MBL) was carried out by the imipenem-ethylenediaminetetraacetic acid disc diffusion/double-disc synergy test. AmpC enzyme production was tested by a modified three-dimensional extract test.
Results:
Almost all isolates were found sensitive to colistin. A high percentage of drug resistance was observed in these isolates against ceftazidime (100%), cefotaxime (100%), cefepime (100%), ofloxacin (97.56%), amoxicillin/clavulanic acid (97.56%) and norfloxacin (85.36%). Of the 41 isolates, ESBL producers were found to be predominant, i.e., 22 (53.65%), followed by AmpC (6, 14.63%) and MBL (5, 12.19%).
Interpretation & conclusions:
At 60 per cent similarity cut-off value, the dendrogram analysis showed that there were a total of 14 unique clusters of ERIC (CL-1 - CL-14) within the 41 E. coli isolates, which revealed the genetic diversity existing between them.
Keywords
β-lactamase
enterobacterial repetitive intergenic consensus polymerase chain reaction
Escherichia coli
multidrug resistance
Escherichia coli is well known to be a universal commensal flora in humans as well as in several animal species but is also reported to be one of the most common enterobacterial species which causes extra-intestinal infections in these hosts1. Clinicians often recommend β-lactam group of antibiotics as an empirical treatment of infections due to E. coli2. The members of Enterobacteriaceae family are mostly associated with the hospital-acquired infections such as gastrointestinal, urinary tract and pyogenic lesions, of which the commonly isolated species is the E. coli3. Increasing prevalence of multidrug-resistant E. coli has been a global public health issue that brings in undesirable hindrance in the diagnosis of infecting agents and treatment options4.
The frequent occurrence of high-level antibiotic resistance is through the transfer of large plasmids which can acquire numerous genes for multiple β-lactamase resistance from different classes of bacteria5. β-lactamase production by several Gram- negative and Gram-positive organisms is perhaps the most important single mechanism of resistance to β-lactam agents3. In addition, resistance to broad-spectrum β-lactams, mediated by extended spectrum β-lactamases (ESBL), metallo β-lactamases (MBL) and AmpC β-lactamases (AmpC) enzymes has been rising problem worldwide6 and has become a major concern as they bring about limitations in therapeutic options, result in treatment failure and are fast propagating throughout the world7.
Strain typing is an epidemiologically important tool not only for detecting the cross transmission of nosocomial pathogens but also for determining the source of infection8. Available sub-typing methods for E. coli include pulsed-field gel electrophoresis (PFGE), plasmid profiling, ribotyping and polymerase chain reaction (PCR)-based typing methods such as arbitrary primed PCR, repetitive extragenic palindromes and enterobacterial repetitive intergenic consensus (ERIC)9. However, the simplicity, ease of operation, low per isolate cost and limited need for specialized laboratory equipment favours the use of ERIC-PCR over other methods10. The present study was carried out to determine the genetic diversity of different β-lactamase producing multidrug-resistant (MDR) E. coli isolates using ERIC-PCR in Odisha in a hospital setup.
Material & Methods
The antibiotics used in this study were purchased from Hi-media, Mumbai. Molecular biology reagents were purchased from Thermo Scientific, India. All other chemicals and reagents were of analytical grade purchased from Hi-media. Thermal cycler and gel documentation system were used for the phylogenetic analyses of the isolates were from Bio-Rad, India.
The present study was carried out from June 2015 to February 2016 in the department of Microbiology, IMS & SUM Hospital, Bhubaneswar, Odisha. During the study period, a total of 103 non-repeated E. coli isolates were obtained from various clinical samples (pus, wound, urine, blood, tracheal aspirate). All isolates were identified morphologically and biochemically by standard procedures. Of the 103 E. coli isolates, 41 (39.8%) were selected for this study as these isolates were multidrug resistant. Details of the sample collection and their processing are presented in the flow chart (Fig. 1).
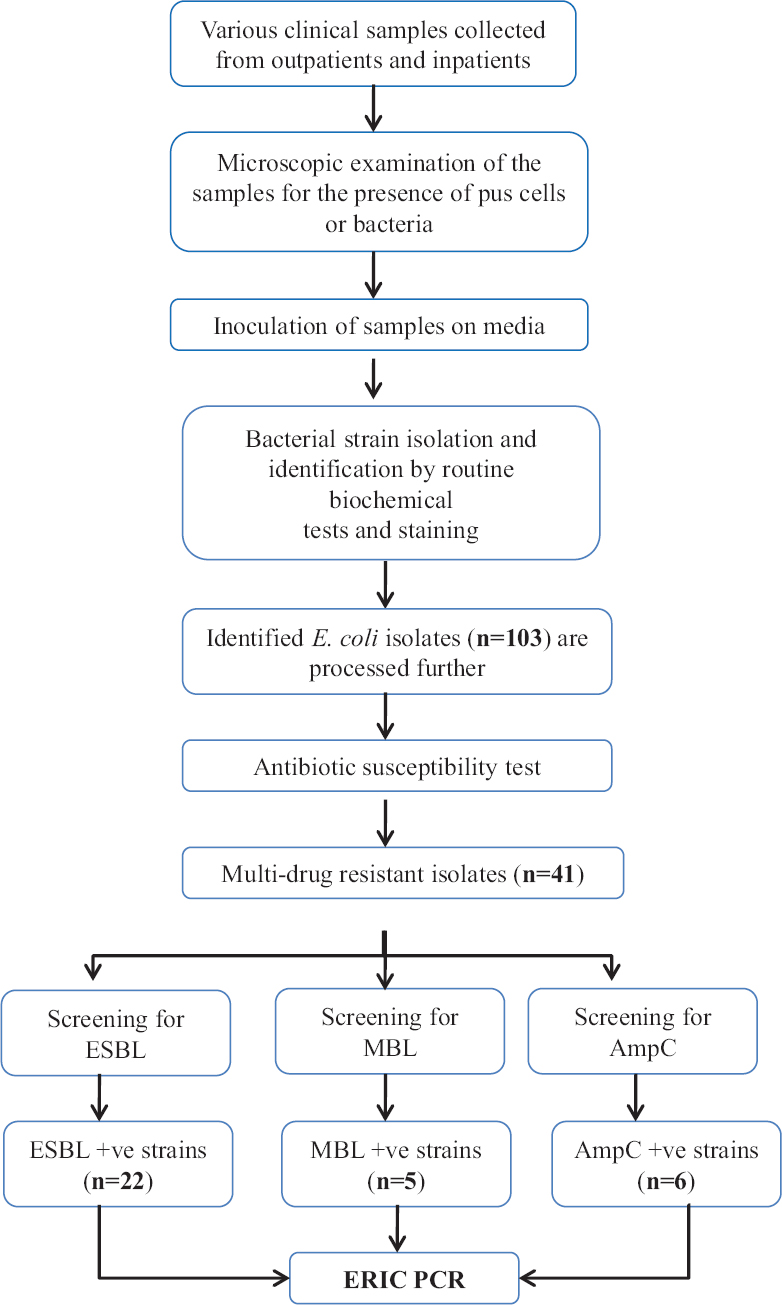
- Flow chart showing study design. ESBL: Extended spectrum β-lactamase, MBL: Metallo β-lactamase, ERIC-PCR: Enterobacterial repetitive intergenic consensus polymerase chain reaction, AmpC: Class C Cephalosporinase.
Antimicrobial susceptibility testing: By using Kirby-Bauer’s disc diffusion method as per Clinical Laboratory Standards Institute (CLSI) guidelines, antimicrobial susceptibility was determined11. Antibiotics used were ceftazidime (30 μg) (CAZ), cefotaxime (30 μg) (CTX), cefepime (30 μg) (CFM) (Cephalosporin), piperacillin/tazobactam (100/10 μg) (PIT), amoxicillin/clavulanic acid (30/10 μg) (AMC) (Penicillin combinations), cefoperazone/sulbactam (75/30 μg) (CFS), ceftazidime/clavulanic acid (30/10 μg) (CAC) (Cephalosporin combinations), ofloxacin (5 μg) (OF), norfloxacin (5 μg) (NX) (Quinolones/fluoroquinolones), amikacin (30 μg), gentamicin (10 μg) (Aminoglycosides), co-trimoxazole (25 μg) (COT) (Sulphonamides), nitrofurantoin (300 μg) (NIT) (Nitrofurans), imipenem (10 μg) (IPM), meropenem (10 μg) (MRP) (Carbapenem), colistin (10 μg) (CL) (Polypeptides) and tigecycline (15 μg) (TGC) (Others). However, COT (25 μg) and NIT (300 μg) were used only in case of urine samples.
Screening for metallo β-lactamases (MBL) production: MBL producing E. coli strains were suspected when the isolates were resistant to IPM/MRP. Phenotypic detection of MBL was carried out by the IPM-ethylenediaminetetraacetic acid disc diffusion/ double-disc synergy test as described previously12.
Detection of extended spectrum β-lactamases (ESBL) & AmpC β-lactamase: Bacteria with resistance or with decreased susceptibility (intermediate by CLSI criteria) to the third generation cephalosporins were tested for ESBL production by NCCLS confirmatory test as described previously12. AmpC enzyme production was tested by a modified three-dimensional extract test as described previously12.
Enterobacterial repetitive intergenic consensus polymerase chain reaction: ERIC-PCR13 was carried out to obtain DNA fingerprints of the E. coli isolates. Colony PCR was carried out using the primers ERIC-IR: 5’-ATGTAAGCTCCTGGGGAATCAC-3’ and ERIC-2: 5’-AAGTAAGTGACTGGGGTGAGCG-3’14. A 25 µl of the reaction mixture was prepared by taking 12.5 µl double-distilled sterile water, bacterial colony (template DNA), 3.0 mM MgCl2 and 200 lm dNTP (each), 0.5 lm primer and 1.0 unit of Taq Polymerase (Merck, Germany). The reaction mixture was initially denatured for three minutes at 94°C, subjected to 40 cycles of denaturation at 94°C for one minute, annealed at 48°C for one minute, extended at 72°C for two minutes, finally extended at 72°C for 10 min and finally soaked to 4°C. The amplified PCR products were analyzed using 1.5 per cent agarose gel electrophoresis. Gel was stained with ethidium bromide and visualized by ultraviolet transilluminator.
Results
MDR E. coli isolates showing resistance to two or more groups of antibiotics and producing any one of the β-lactamases (ESBL, MBL and AmpC) were included. The majority of isolates (30: 73.17%) were from urine samples, and the rest were from other body fluids.
The antibiotic sensitivity pattern of the MDR E. coli isolates revealed that the maximum resistance was observed against CAZ (100%), CTX (100%), CFM (100%), OF (97.56%), AMC (97.56%) and NX (85.36%). Lower resistivity to a range of 39-54 per cent, was observed in β-lactamase inhibitors CFS, CAC and PIT except AMC. The lowest rate of resistance was observed in TGC (7.31%), IPM (17%), MRP (26.82%) and NIT (22%) (data not shown). It was found that all the MDR β- lactamase producing E. coli isolates are sensitive to CL. Among all the β-lactamases, ESBL were found to be higher in number, i.e., 22 (53.65%), followed by AmpC 6 (14.63%) and MBL 5 (12.19%) (Fig. 2).
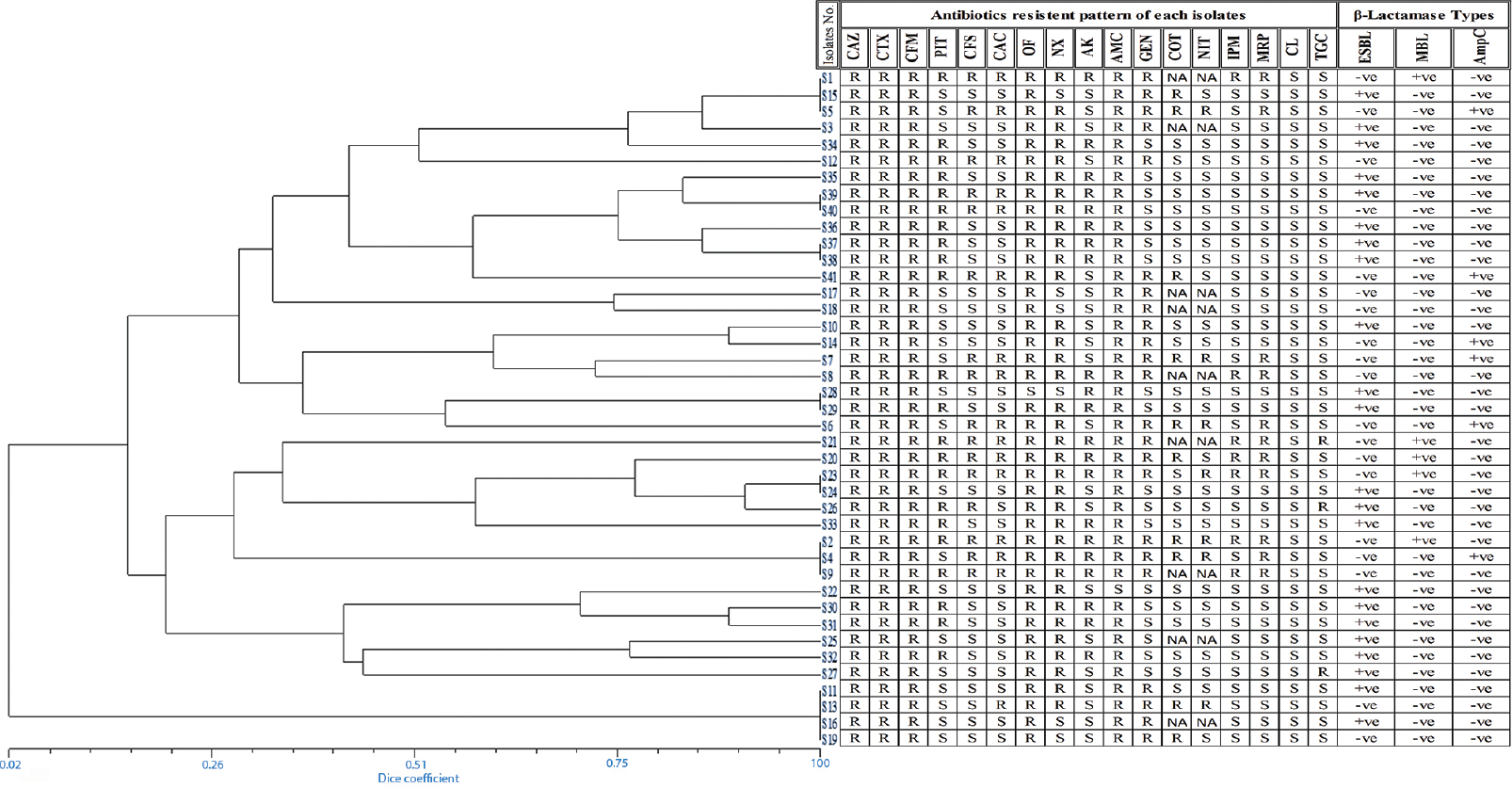
- Phylogenetic relationships among Escherichia coli isolates generated using UPGMA based on dice coefficients method derived from analysis of the ERIC-PCR profiles. Antibiotic resistance profile and β-lactamase type are also shown. R: Resistant, S: Sensitive, NA: Not applied, +ve: Positive, −ve: Negative.
The genetic diversity analysis of 41 non-repeated randomly collected β-lactamase-producing E. coli isolates was carried out by ERIC-PCR fingerprint method using ERIC-IR and ERIC-2 primers. The fingerprints obtained from the ERIC typing of the 41 MDR E. coli isolates showed a DNA banding profile consisting of amplified bands ranging from 1 to 7 having size 150 bp to 2500 bp (Fig. 3A & B). At 60 per cent similarity cut-off value of the dendrogram analysis showed that there were a total of 14 unique clusters of ERIC (CL-1 to CL-14) within the 41 E. coli isolates, exhibiting genetic diversity (Fig. 4). Only five clusters (CL-1, CL-2, CL-6, CL-8 and CL-14) represented four or more isolates while the remaining nine clusters contained less number of isolates varying from 1 to 3. Some isolates showing similar ERIC profile, four from CL-1 (S19, S16, S11 and S13), three from CL-8 (S15, S1 and S5) and three from CL-12 (S9, S2 and S4), which indicated clonal similarities between the isolates.
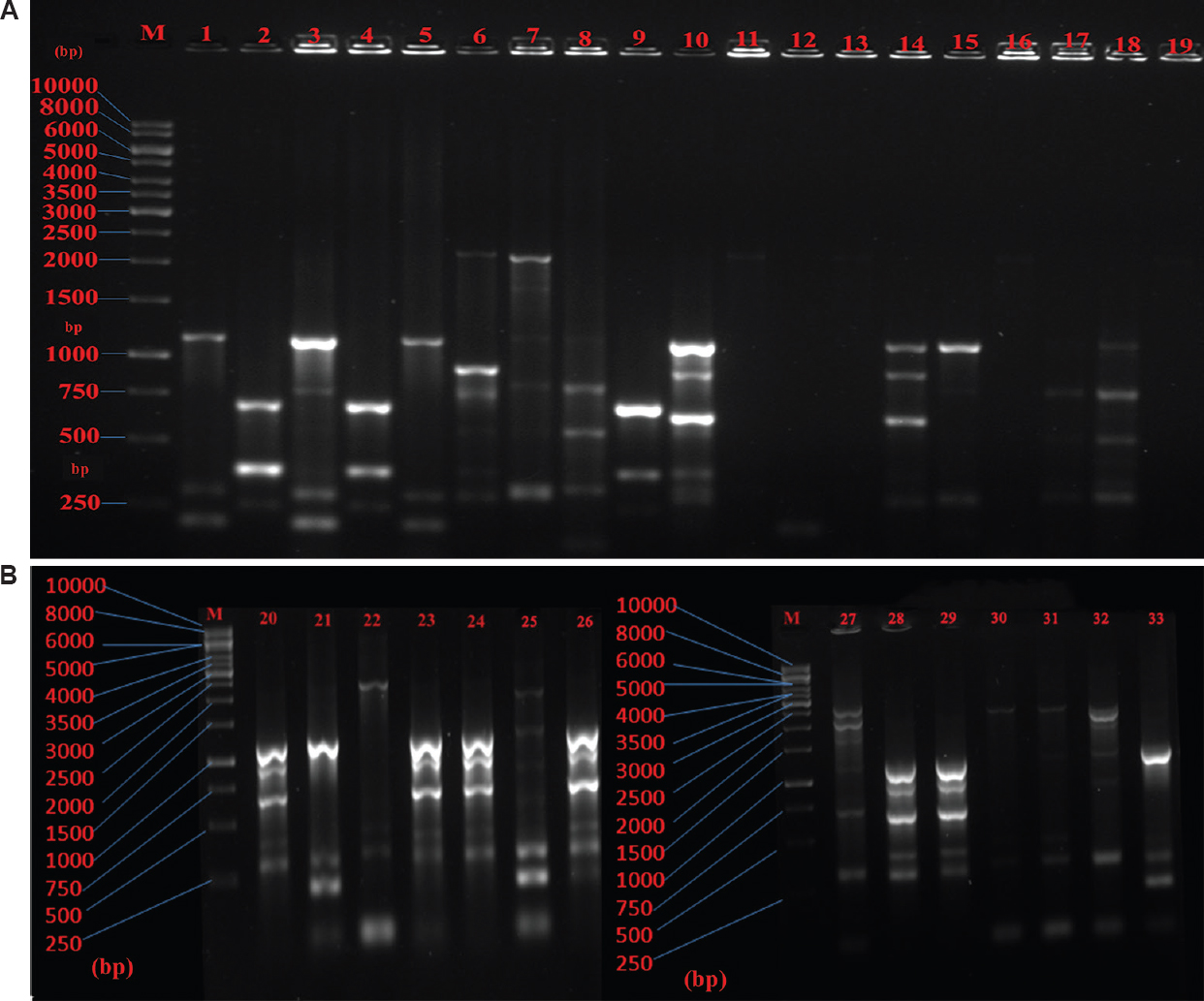
- (A) Agarose gel electrophoresis of enterobacterial repetitive intergenic consensus polymerase chain reaction (ERIC-PCR) generated DNA fingerprints of 1-19 Escherichia coli isolates. M: Marker (1 kb), (B) Agarose gel electrophoresis of ERIC-PCR generated DNA fingerprints of 20-33 Escherichia coli isolates. M: Marker (1 kb).
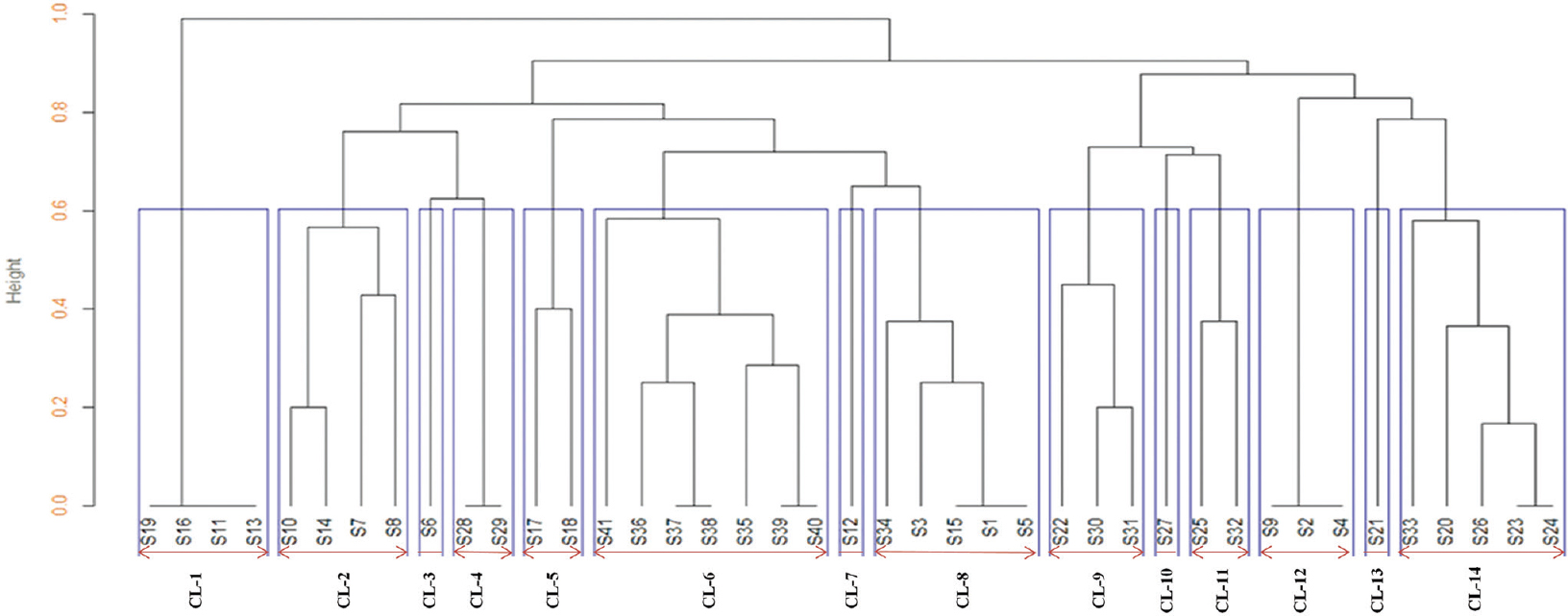
- Dendrogram of Escherichia coli isolates at 60 per cent similarity cut-off value analysis, it showed 14 unique clusters. CL: Cluster.
Discussion
The development of MDR due to the production of β-lactamases in Gram-negative bacteria has become a major problem worldwide, which limits the treatment option and increases the hospital cost, the rate of morbidity and mortality. The sources of infection and spreading pattern may generally be understood by analyzing the clonal similarity among the isolates collected from any hospital setup using specific molecular marker.
In the present study, the maximum number of isolates (73.17%) were from urinary tract infections (UTI) which corroborated with several other studies described previously151617. The reason being E. coli, the most prevalent causative organisms of UTI is solely responsible organism for more than 80 per cent of the infections18.
In our study, the rate of resistance to CAZ (100%) and AMC (97.56%) were much higher when compared with other studies from Malaysia19 and China where it was found to be 28 per cent for CAZ and 84 per cent AMC20. CFM showed 100 per cent resistance in our study, which was much higher when compared with another study19 reporting only two per cent. Carbapenem group of antibiotics, such as IPM and MRP showed resistance at 17 and 26.82 per cent, respectively, in our study, but Adwan et al21 showed the rate of resistance to IPM to be 22 per cent in E. coli and obtained no MRP resistance isolates. A study by Rezai et al22 reported high CL resistant (82%) in ESBL-producing uropathogenic E. coli. However, the present study showed complete (100%) susceptibility to CL.
The prevalence of ESBL-producing E. coli varies from country to country and from centre to centre. Our study showed the prevalence of ESBL was 53.65 per cent, which was higher than the study from Sudan with 30.2 per cent ESBL producers in E. coli23. It has been shown that the prevalence of ESBL-producing E. coli is highest in India (60%), followed by Hong Kong (48%) and Singapore (33%)24. The prevalence of ESBL-producing isolates of E. coli was reported to be 13.3 per cent in Lebanon25, 9.2 per cent in Korea26, 10.3 per cent in Arabia27 and 17 per cent in Turkey28, which was much low in prevalence when compared to India2930. In the present study, we found 14.63% AmpC β-lactamase producers which were lower when compared with the studies from Chennai (in children under five) and Kolkata (37.5 and 47.8 per cent, respectively)3132. We found 12.19 per cent were MBL producers, which was lower as compared with the study by Bora et al33 from Nepal with a higher incidence of MBL-producing E. coli (18.98%) from different clinical samples.
In our study 14 unique clusters of ERIC were observed in 41 MDR E. coli isolates at a 60 per cent cut-off value analysis and exhibited genetic heterogeneity, but Adwan et al21 reported 11 unique clusters of ERIC in 41 MDR E. coli which showed genetically diverse.
On the basis of ERIC-PCR fingerprint, the isolates included in our study were genetically diverse. This is the most expected outcome as the isolates were randomly collected from different sources and from different samples, which demonstrate that, the transmission might have occurred from clones of different origin. These findings corroborated with other studies213435.
In the present study, some isolates had similar ERIC profile, four from CL-1 (S19, S16, S11 and S13), three from CL-8 (S15, S1 and S5) and three from CL-12 (S9, S2 and S4) showed genetically homogeneous profile. Similar report was given by Loncaric et al36 analyzing bacterial isolates from migratory and resident population of Rooks birds (Corvus frugilegus) from Austria. From this study, we observed that there is no correlation of the clonally similar strains with their respective antibiotic sensitivity nor with pattern the β-lactamase enzymes they produced. Loncaric et al36 demonstrated that the spread of antibiotic resistance not only due to the dissemination of different β-lactamases but also due to clonal transmission.
In the present investigation, only a few isolates showed conserved banding profile on the basis of ERIC analyses, which indicated the rapid dissemination of similar clonal groups. A similar report was given by Durmaz et al34 on ESBL-producing quinolone-resistant clinical E. coli isolates. Shakil et al37 reported clonal similarity among four blaCTX-M-15-positive E. coli isolates from NICU patients by ERIC-PCR and assigned them to be temporarily responsible for clonal outbreak. From the present study, it can be concluded that ERIC-PCR-based strain typing of β-lactamase producing MDR E. coli collected from various clinical samples exhibited a considerable amount of clonal heterogeneity. The occurrence of a few clonally similar groups among these isolates could be either due to the spread of antibiotic resistance through dissemination of different β-lactamases or due to clonal transmission. However, lack of association of these clonally similar isolates with their respective antibiotic sensitivity pattern or with the β-lactamase enzyme production in our study rules out above possibility, indicating antibiotic selection pressure to be one of the major reasons which need to be validated further.
The sample size of the present study was small. Multicentric study with larger sample size, however, may help better in establishing the genetic diversity existing among the E. coli isolates in a region.
Acknowledgment
Authors acknowledge Dr D.K. Roy, Dean, IMS & SUM Hospital (S‘O’A University) for extended essential laboratory facilities and Dr M. R. Nayak, Honourable President, Siksha ‘O’ Anusandhan University, Bhubaneswar, for providing financial support.
Conflicts of Interest: None.
References
- Extended-spectrum beta-lactamases: A clinical update. Clin Microbiol Rev. 2005;18:657-86.
- [Google Scholar]
- Prevalence of extended-spectrum beta-lactamase and class 1 integron integrase gene intl1 in Escherichia coli from Thai patients and healthy adults. Southeast Asian J Trop Med Public Health. 2008;39:425-33.
- [Google Scholar]
- Extended spectrum -lactamases (ESBL) –An emerging threat to clinical therapeutics. Indian J Med Microbiol. 2004;22:75-80.
- [Google Scholar]
- First description of SHV-148 mediated extended-spectrum cephalosporin resistance among clinical isolates of Escherichia coli from India. Indian J Med Microbiol. 2016;34:33-7.
- [Google Scholar]
- Phenotypic and genotypic characterization of Enterobacteriaceae with decreased susceptibility to carbapenems: Results from large hospital-based surveillance studies in China. Antimicrob Agents Chemother. 2010;54:573-7.
- [Google Scholar]
- Evaluation of methods for AmpC beta-lactamase in gram negative clinical isolates from tertiary care hospitals. Indian J Med Microbiol. 2005;23:120-4.
- [Google Scholar]
- Detection of Amp C genes encoding for beta-lactamases in Escherichia coli and Klebsiella pneumoniae. Indian J Med Microbiol. 2012;30:290-5.
- [Google Scholar]
- Phylogenetic study of metallo-β-lactamase producing multidrug resistant Pseudomonas aeruginosa isolates from burn patients at a tertiary care Hospital. Burns. 2015;41:1758-63.
- [Google Scholar]
- Characterisation and genetic diversity via REP-PCR of Escherichia coli isolates from polluted waters in Southern Brazil. FEMS Microbiol Ecol. 2003;45:173-80.
- [Google Scholar]
- Facilitated molecular typing of Shigella isolates using ERIC-PCR. Am J Trop Med Hyg. 2012;86:1018-25.
- [Google Scholar]
- Performance Standards for Antimicrobial Susceptibility Testing;Twenty-Fourth Informational Supplements M100-S24. Wayne, PA: CLSI; 2014.
- Prevalence of ESBL, MBL and AmpC β-lactamases producing multidrug resistance gram negative bacteria in a tertiary care hospital. J Pure Appl Microbiol. 2014;8:4099-105.
- [Google Scholar]
- Biodegradation and bioremediation of endosulfan contaminated soil. Bioresour Technol. 2008;99:3116-22.
- [Google Scholar]
- Distribution of repetitive DNA sequences in eubacteria and application to fingerprinting of bacterial genomes. Nucleic Acids Res. 1991;19:6823-31.
- [Google Scholar]
- High prevalence of extended-spectrum b-lactamase-producing pathogens: Results of a surveillance study in two hospitals in Ujjain, India. Infect Drug Resist. 2012;5:65-73.
- [Google Scholar]
- ESBL, MBL and Ampc b Lactamases producing superbugs –Havoc in the Intensive Care Units of Punjab India. J Clin Diagn Res. 2013;7:70-3.
- [Google Scholar]
- Antibiotic resistance and genotype of beta-lactamase producing Escherichia coli in nosocomial infections in Cotonou, Benin. Ann Clin Microbiol Antimicrob. 2015;14:5.
- [Google Scholar]
- Outcome of cephalosporin treatment for serious infections due to apparently susceptible organisms producing extended-spectrum β-lactamase: Implications for the clinical microbiology laboratory. J Clin Microbiol. 2001;39:2206-12.
- [Google Scholar]
- Characterization of multidrug resistant ESBL-producing Escherichia coli isolates from hospitals in Malaysia. J Biomed Biotechnol 2009 2009:165637.
- [Google Scholar]
- Resistance of strains producing extended-spectrum beta-lactamases and genotype distribution in China. J Infect. 2007;54:53-7.
- [Google Scholar]
- Molecular characterization of Escherichia coli isolates from patients with urinary tract infections in Palestine. J Med Microbiol. 2014;63(Pt 2):229-34.
- [Google Scholar]
- Characterization of multidrug resistant extended-spectrum beta-lactamase-producing Escherichia coli among uropathogens of pediatrics in North of Iran. Biomed Res Int 2015 2015:309478.
- [Google Scholar]
- Prevalence of extended-spectrum b-lactamases-producing Escherichia coli from Hospitals in Khartoum State, Sudan. Oman Med J. 2013;28:116-20.
- [Google Scholar]
- Consensus review of the epidemiology and appropriate antimicrobial therapy of complicated urinary tract infections in Asia-Pacific region. J Infect. 2011;63:114-23.
- [Google Scholar]
- Prevalence of the genes encoding extended-spectrum beta-lactamases, in Escherichia coli resistant to beta-lactam and non-beta-lactam antibiotics. Ann Trop Med Parasitol. 2005;99:413-7.
- [Google Scholar]
- Molecular characterization of extended-spectrum beta-lactamases produced by clinical isolates of Klebsiella pneumoniae and Escherichia coli from a Korean nationwide survey. J Clin Microbiol. 2004;42:2902-6.
- [Google Scholar]
- Prevalence and antimicrobial susceptibility of extended-spectrum beta-lactamase-producing Escherichia coli and Klebsiella pneumoniae in a general hospital. Ann Saudi Med. 2005;25:239-42.
- [Google Scholar]
- Extended-spectrum β-lactamases among cloacal Escherichia coli isolates in healthy broilers in Turkey Turk. J Vet Anim Sci. 2017;41:72-76.
- [Google Scholar]
- Cefoxitin resistance mediated by loss of a porin in clinical strains of Klebsiella pneumoniae and Escherichia coli. Indian J Med Microbiol. 2005;23:20-3.
- [Google Scholar]
- Urinary tract infection and antimicrobial susceptibility pattern of extended spectrum of β-lactamase producing clinical isolates. Biol Res. 2008;2:78-82.
- [Google Scholar]
- AmpC beta-lactamase producing multidrug resistant strains of Klebsiella spp. & Escherichia coli isolated from children under five in Chennai. Indian J Med Res. 2003;117:13-8.
- [Google Scholar]
- AmpC β-lactamase producing bacterial isolates form Kolkata hospital. Indian J Med Res. 2005;122:224-33.
- [Google Scholar]
- Incidence of metallo-beta-lactamase producing clinical isolates of Escherichia coli and Klebsiella pneumoniae in central Nepal. BMC Res Notes. 2014;7:557.
- [Google Scholar]
- Detection of β-lactamase genes, ERIC-PCR typing and phylogenetic groups of ESBL producing quinolone resistant clinical Escherichia coli isolates. Biomed Res. 2015;26:43-50.
- [Google Scholar]
- Emergence of integron borne PER-1 mediated extended spectrum cephalosporin resistance among nosocomial isolates of Gram-negative bacilli. Indian J Med Res. 2015;141:816-22.
- [Google Scholar]
- Comparison of ESBL –and AmpC producing Enterobacteriaceae and methicillin-resistant Staphylococcus aureus (MRSA) isolated from migratory and resident population of rooks (Corvus frugilegus) in Austria. PLoS One. 2013;8:e84048.
- [Google Scholar]
- Acquisition of extended-spectrum beta-lactamase producing Escherichia coli strains in male and female infants admitted to a neonatal Intensive Care Unit: Molecular epidemiology and analysis of risk factors. J Med Microbiol. 2010;59(Pt 8):948-54.
- [Google Scholar]






