Translate this page into:
Genetic architecture of HIV-1 genes circulating in north India & their functional implications
Reprint requests: Dr Akhil C. Banerjea, Chief & Staff Scientist VII, Laboratory of Virology, National Institute of Immunology, JNU Campus, Aruna Asaf Ali Marg, New Delhi 110 067, India e-mails: akhil@nii.res.in; akhil_banerjea@yahoo.com
-
Received: ,
This is an open-access article distributed under the terms of the Creative Commons Attribution-Noncommercial-Share Alike 3.0 Unported, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
This article was originally published by Medknow Publications & Media Pvt Ltd and was migrated to Scientific Scholar after the change of Publisher.
Abstract
This review presents data on genetic and functional analysis of some of the HIV-1 genes derived from HIV-1 infected individuals from north India (Delhi, Punjab and Chandigarh). We found evidence of novel B/C recombinants in HIV-1 LTR region showing relatedness to China/Mynmar with 3 copies of Nfκb sites; B/C/D mosaic genomes for HIV-1 Vpr and novel B/C Tat. We reported appearance of a complex recombinant form CRF_02AG of HIV-1 envelope sequences which is predominantly found in Central/Western Africa. Also one Indian HIV-1 envelope subtype C sequence suggested exclusive CXCR4 co-receptor usage. This extensive recombination, which is observed in about 10 per cent HIV-1 infected individuals in the Vpr genes, resulted in remarkably altered functions when compared with prototype subtype B Vpr. The Vpu C was found to be more potent in causing apoptosis when compared with Vpu B when analyzed for subG1 DNA content. The functional implications of these changes as well as in other genes of HIV-1 are discussed in detail with possible implications for subtype-specific pathogenesis highlighted.
Keywords
HIV-1 genetic variants
HIV-1 recombinants
HIV-1 subtypes
India
Introduction
Since the first detection of HIV-1 in India1 in Chennai, Tamil Nadu, it has now been reported from almost all the s0 tates of India. The prevalence rates vary from 0.2-0.3 per cent in majority of the States to almost about 1 per cent reported for two southern s0 tates. The reasons for this discrepancy are not known and remain an interesting research problem. The HIV-1 epidemic in India started with genetic subtype C which probably entered through Southern Africa and it is believed that even today it is the most predominant subtype in all the s0 tates. The current estimate of HIV-1 infected individuals in India is 2.5 to 3.1 million based on the surveillance report of National AIDS Control Organization (NACO) and other agencies2. The molecular epidemiology of HIV-1, drug resistance, immune response and host genetics with reference to Indian scenario have been reviewed by several Indian investigators3–6 including our group78. This mostly covers the HIV-1/AIDS scenario based on studies carried out mostly in southern India. Most of these studies were restricted to genetic analyses and very little information was available with respect to functional implications of the genetic changes in the viral genes.
India is a large country and independent pockets of HIV-1 infections may be forming at multiple regions of India. Since very little information on the nature of HIV-1 epidemic in north India is available, we will focus, in this review, the genetic architecture of HIV-1 genes circulating in this region and functional implications of some of the genes which may help in understanding the molecular basis of genetic subtype-specific pathogenesis. We report the novel mosaic/ recombinant structures of several important HIV-1 genes; increasing frequency of B/C or B/C/D genome segments and other novel and complex recombination events in the open reading frames (ORFs) of several viral genes which underscores the importance of co-circulation of multiple genetic subtypes that are likely to impact various HIV-1 prevention programmes. These recombinants are unique in the sense that cross-over events have taken place within the short ORFs of several HIV-1 genes. Some of these recombinants have widely divergent functions. We have deposited approximately 200 HIV-1 gene sequences from north India in the data bank and can be accessed from www.pubmed.gov site (select nucleotide from the menu and type “banerjea a” - to retrieve sequences - most of them are named as NII-PGI-viral genes).
Global HIV-1 epidemiology and Indian scenario
HIV diversity is associated with the error prone reverse transcriptase enzyme. While subtype A is prevalent in Africa, subtype B is predominant in North and South American continent. The Indian subcontinent has subtype C as the major prevalent subtype along with China and South Africa, which is the major subtype contributing to the epidemic worldwide. Although, there is no direct correlation between the fitness of various subtypes to their respective spread, the prevalence of subtype C among majority of the HIV infected individuals throughout the world would suggest that it has been naturally selected over other subtypes. The transmission rate of subtype C HIV-1 long terminal repeats (LTRs) from mother to infant was shown to be more efficient when compared to either subtype A or inter-subtype recombinants9. Also, in a recent review by scientists at National AIDS Research Institute, Pune, suggested that subtype C possessed fitness advantage over subtype A3. Most of the studies pertaining to HIV-1 pathogenesis have been carried out on subtype B from Europe and USA. In contrast little information is available for subtype C, responsible for majority of the global epidemic (Africa and Asia). It has been suggested that the most recent ancestor of genetic subtype C appeared in Karonga District in Malawi in the mid-to late 196010. Numerous reports suggest the presence of subtype C throughout India11–13. Less prevalent HIV-1 subtypes, other than subtype C, have also been reported by several investigators in different parts of the country. These include subtypes B and A from New Delhi and Punjab (northern India), West Bengal and Manipur (eastern and north-eastern region), Maharashtra, Andhra Pradesh, Tamil Nadu and Gujarat (southern, western, and south western)14–21. So, it would be worthwhile determining the implications of other subtypes on HIV pathogenesis and their impact on the spatio-dynamics of HIV-1 epidemic.
HIV-1 LTR promoter variants, transactivation potential and mother-to-child transmission
HIV-1 LTR promoter region governs HIV-1 transcription and contains several transcription factor binding sites. Out of several transcription factors, NfkB has attracted the most attention. Most subtype B HIV-1 LTRs possess 2 whereas LTR C is known to possess 3 or sometimes even 4 copies of it which often positively correlates with HIV-1 gene expression. The most intriguing feature is the conservation of short subtype-specific sequences between the two successive Nfkb sites. We recently reported novel HIV-1 LTR B/C recombinants that had segments derived from B/C China/Mynamar, B/C India22 (Fig. 1). There were numerous changes in the nucleotide sequences of the LTR region (~ 650 bases) which include the transcription factor binding regions. These novel mosaic B/C LTRs retained their ability to get activated by HIV-1 Tat protein. We have further extended this study and characterized 4 pairs of mother-child samples with respect to conservation of LTR sequences as well as their ability to get transactivated with HIV-1 Tat protein. Three out of four LTR sequences from children samples exhibited increased transactivation when compared with their mother's LTR sequences (unpublished observation) and this observation closely mimics the observation made for subtype B LTR sequences carried out in USA23. Thus high expression LTR phenotypes are selectively transmitted.
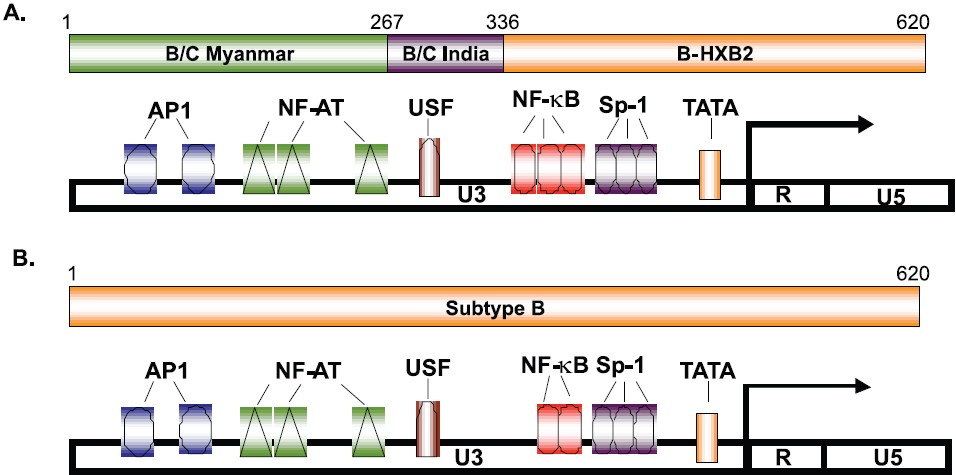
- Mosaic pattern and organization of transcription factor binding sites in HIV-1 LTR reported from north India: (A) B/C LTR showing 3 NF-κB (B) B LTR showing 2 NF-κB. Source: Ref. 22.
HIV-1 Tat variants - presence of a novel Tat B/C recombinant
HIV-1 Tat is a multifunctional protein that is intricately involved with HIV-1 gene expression and replication. It is produced from multiply spliced HIV-1 transcript and usually the 1st exon is sufficient for majority of its known functions24. It consists of at least 4 well defined domains. It is noteworthy that the cysteine rich domain of subtype C possesses C31S mutation. This has enormous implications on its chemokine activity which is linked to very low incidence of HIV-associated dementia (HAD) in subtype C infected Indian patients as reported earlier25. Subsequent studies from the National Brain Research Institute, Manesar, Haryana, using human foetal central nervous system progenitor cell-derived astrocytes and neurons found clade B Tat protein or DNA to be more potent as compared to those of clade C in inducing apoptosis in the neurons and successfully demonstrated clade-specific functional differences26. It is also worth noting that most subtype C isolates possess QGD motif in the second exon of Tat but subtype B isolates possess RGD motif at similar position. Our group at National Institute of Immunology, New Delhi, created chimeric Tat constructs possessing domains from subtype B and C (from an Indian isolate 93IN905). This study clearly indicated that subtype B was more apoptogenic for T and monocytic cells. It also established that QGD motif present in subtype C Tat increased the Tat B mediated transactivation 3-fold more27. When the Tat gene was analyzed from the infected individuals, we occasionally observed (3 out of 30) novel B/C recombinant (sample A9; Fig. 2). A precise breakpoint was observed at nucleotide position 123- the N-terminal half consisted of subtype B and the C-terminal of subtype C (unpublished observation). We observed that subtype B Tat was always more potent activator of LTR promoter than C Tat. When this recombinant Tat gene was tested for its ability to activate HIV-1 LTR promoter, it activated with intermediate efficiency. LTR activation related activities of Tat variants correlated with their ability to bind to Tar RNA (unpublished data). Potent anti-Tat antibody production has been reported in some infected individuals which could neutralize a wide variety of genetic subtypes28. Since Tat plays a very dominant role in HIV-1 gene expression and replication, the appearance of natural mutations and their functional implications will constitute an important comprehensive epidemiological study.
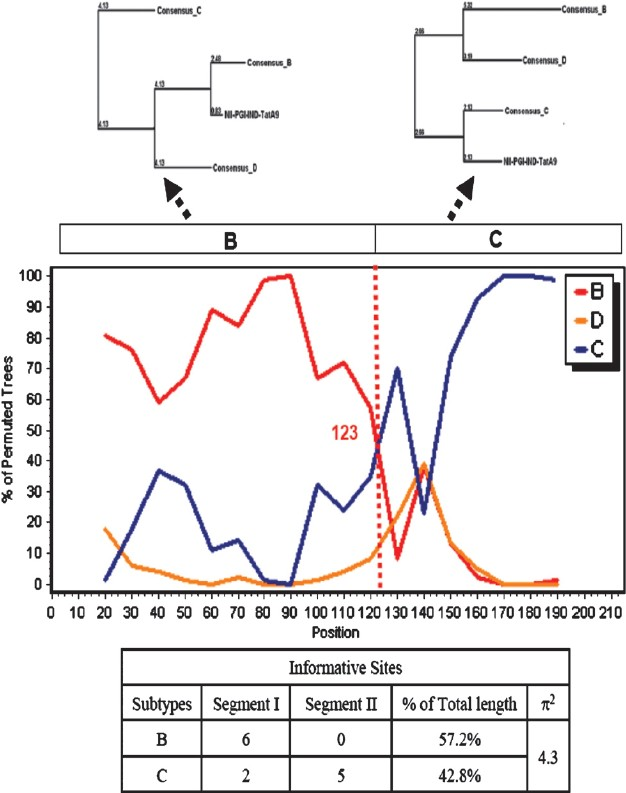
- Bootscan, informative and phylogenetic analysis of NIIPGI- IND-TatA9 variant using SimPlot ver 3.5.1. Bootscan analysis was carried out using Kimura (2-parameter) with a window size of 40 bp and a stepsize of 20 bp. Precise breakpoint and genomic makeup of the recombinant A9 isolate is shown.
Sequence variation and functional domain analysis of Vif B and C genes
Vif is an accessory protein originally described as Virion infectivity factor. It is present in almost all lentiviruses and is required for viral replication and pathogenesis in vivo29. It is mainly involved in neutralizing the DNA-editing enzymes, namely, cytidine deaminases of APOBEC family of proteins by promoting their degradation by proteasomal pathway. There are very significant diffrences in the sequence of aminoacids of Vif derived from a prototypic subtype B (pNL4-3) and a prototypic subtype C derived from an Indian isolate -93IN905- originally described by Lole et al30. This subtype C isolate is very close (>90%) to the consensus subtype C and, therefore, extensively used as a standard by various investigators. Gupta & Banerjea31 made several chimeric B/C mutants of this protein and broadly defined the domains responsible for APOBEC protein degradation. Their study suggested that C-terminal of Vif C possessed the major determinants for this function. We analyzed the Vif sequences derived from north Indian population. About 10 per cent of the samples exhibited a novel B/C recombinant structure with a precise break-point at position 292. The N-terminal half was derived from B subtype and the C-terminal half from C subtype. This Vif B/C recombinant possessed ability to degrade APOBEC protein comparable to Vif C (unpublished data). This is the first novel B/C recombinant from India with a precise breakpoint in the middle of the ORF of this gene (data not shown).
Novel B/C/D mosaic structure of Vpr genes - differential apoptotic and transactivation potential
HIV-1 Vpr, another viral accessory gene plays an important role in virus replication and can influence several viral and cellular functions (reverse-transcription process, nuclear import of viral DNA, cell cycle arrest, activation of cellular and HIV-1 LTR promoter32. Therefore, genetic and functional analysis of the Vpr gene from eight HIV-1 infected individual from north India was recently carried out by our group to understand its possible role in HIV-1/AIDS pathogenesis33. Vpr gene from six out of the eight HIV-1 infected samples showed clustering with subtype C and possessed L64P mutation. Two samples were recombinants consisting of B, C and D subtype derived genomes (Fig. 3). The most interesting feature between the two recombinants was the presence of either subtype C or subtype B genome segments at their C-terminal halves. Their N-terminal halves consisted of identical mosaic B/C and D structure. Also functional analysis of these six samples showed that two L64P mutants (VprS1 and S3) poorly activated HIV-1 LTR promoter but the recombinant VprS2 was fully competent. All the three samples (VprS1, S2 and S3) retained their ability to cause apoptosis. The VprC variants with L64P mutation show selective functional inactivation. Thus, Vpr recombinants with mosaic genome segments highlight the importance of multiple genetic subtypes in shaping the HIV-1 epidemic in north India. This study also reinforces the fact that 291nt encoding Vpr is a hot spot for the formation of inter-subtype recombinants.
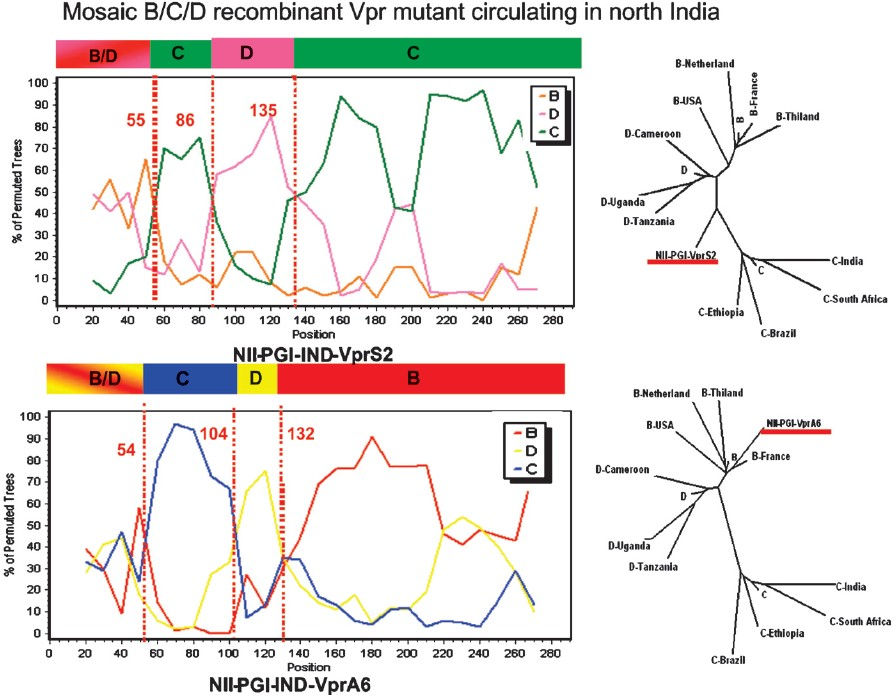
- Genetic analysis of the two Vpr recombinant samples from north India: Similar to Fig. 2, genetic tools were used to characterize the two Vpr sequences. The nature of the mosaic genome segments are shown along with their phylogenetic analysis. Source: Ref. 33.
HIV-1 envelope sequence variants
This gene is undoubtedly the most rapidly accumulating mutations during the course of the disease. It has been made up of multiple constant and variant regions. The gp41 region possesses the fusogenic region and HIV-1 gp120 possesses the CD4 receptor and co-receptor binding regions. It is very heavily glycosylated and responsible for eliciting the antibody responses. Evidence of mosaicism involving A, E and G in this region was reported as early as in 199930 from southern India. Using prototypic subtype C envelope the roles of V3 and V5 in viral entry were defined34. Two groups3536 reported southern subtype C isolates with novel co-receptor requirements to initiate infection. Our group has reported for the first time appearance of CRF_02AG (Central and western African origin) from Delhi besides subtypes B and C37 (Fig. 4). Remarkably this study also predicted one of the subtype C isolates from northern India to be X4-tropic using two different programmes. This observation is important because earlier published work suggested that subtype C infections do not show R5 to X4 co-receptor (chemoikine receptor) switch observed among majority of subtype B infections38. The remarkable feature of the V3 loop, which is primarily responsible for generating neutralizing antibodies and determining the co-receptor usage, is the presence of subtype-specific GPGQ and GPGR sequences in the V3 crown sequence. Our group39 showed the presence of CCR5 Δ32 in the Indian population and this was an extremely rare event. Thus our population is genetically more prone for HIV-1 infection and progression39. Our group was also the first to exploit this target as an antiviral approach (ribozymes, catalytic DNAs, siRNAs)40. Several other genes modulate HIV-1 progression and have recently been reviewed by us8.

- Radial phylogenetic analysis of the envelope sequences. Note the Indian CRF02_AG clustering with earlier described CRF_02AG strains. The other two clusters are B and C subtype-specific.
Rev Response Element (RRE) RNA and Rev protein interaction
The amino acid sequence of Rev B and Rev C (derived from prototype subtype C – 93IN905) differs considerably in the RNA binding domain and the same is true for the 248nt long sequence of RRE. Rev protein is critically involved in the cytoplasmic transport of full length (~9kb genomic) and singly spliced envelope transcript. We observed that Rev B protein was very efficient in binding either RRE B or RRE C. On the other hand, Rev C showed RRE C binding but it failed to show RRE B binding (unpublished observations). These observations are likely to influence genetic subtype-specific gene expression. It is, therefore, important to monitor the nature and extent of mutations in Rev gene and RRE sequences from the same genomic RNA from clinical isolates. We observed that despite changes in the RRE RNA structure, the Rev binding region was retained by carrying out interaction studies between RRE RNA + purified Rev protein by gel analysis41.
HIV-1 Vpu subtype-specific differences
We have recently reported a novel function of HIV-1 Vpu with respect to stabilization of tumour suppressor p53 which is TrcP dependent. It inhibits the ubiquitination of p53. The stabilization of p53 leads to more apoptosis in human T-cells, a hall mark of HIV-1 pathology42. We observed that Vpu C was more potent in causing apoptosis as measured by SubG1 content42. We have sequenced subtype B and C-specific Vpu from HIV-1 infected individuals from north India and observed novel mutations (deletions in the trans-membrane and more substitutions in the C-terminal of Vpu protein variants (data not shown).
Conclusions
The hallmark of HIV is its widespread genetic diversity. The error prone reverse transcriptase (3X10-5 sites/genome/replication cycle) coupled with its enormous capacity to produce virus in vivo at a rate exceeding 109 per day and its persistent nature of infection, there exists tremendous scope for the generation of viral diversity43. Recombinant viruses have already contributed substantially to the global pandemic, and the likelihood of generating recombinant viruses will continue to increase as the different HIV-1 subtypes spread worldwide44. Mixing of different lineages and clades of HIV-1 strains could quickly lead to the evolution of new recombinant strains. Even recombinant viruses will recombine, evolving to produce the second generation recombinants, termed as inter CRF recombinants (ICRs). As an example a CRF namely ICR01_0708 was identified among intravenous drug users (IDUs) in Yunnan Province of China, composed of two closely related CRFs, CRF07_BC and CRF08_BC co-circulating in China4546. There have been numerous reports for the presence of different subtypes in India in which different viral genes of different subclades exist together to produce a recombinant virus but the presence of sequences belonging to different subtypes within the same ORF of the viral genes has been reported recently by our group for Vpr and LTR genes2333. We have emphasized the contribution of these short stretches of gene sequences in modulating the evolution and spread of virus in this region. Although the L64P mutation in the Vpr gene found in most of the samples analyzed, showed reduction in the transactivation capabilities, these were equally competent to the prototype B and C Vpr in inducing apoptosis. This is not surprising as others have reported similar observations with artificially created mutants in Vpr B genes47. We also suggest that these recombination events within the open reading frame of the viral gene are not random but predictable as the breakpoints relate to the points having low entropy values which correlated with the ability to form secondary structures (mountain plot)33. The virus has to selectively accumulate the functionally active mutations which do not hamper the normal pathogenesis and this hypothesis has been supported by experiments carried out using HIV-1 env gene by different groups4849. Thus, there exists a functional constraint over recombination events occurring within these small gene segments. Recombination may thus significantly accelerate viral diversification and facilitate them to develop multi-drug resistance in infected individuals50–53. But, the emergence of viral variants which carry a novel pattern of mutations, conferring them with resistance against antiretroviral therapy, can lead to a loss of diversity throughout the viral genome, even when only a single gene segment is under selective pressure54. Also, many believe recombination increases human immunodeficiency virus fitness, but not necessarily its diversity55.
The pure HIV subtypes are gradually phasing out and are being replaced by mosaic genome viruses owing to the recombinogenic nature of the virus56–58. Recent studies suggest the co-existence of multiple HIV proviruses in infected cells, which enable the formation of heterozygous virions and presents necessary substrate for recombination which induces genomic diversification5960. Many recombinants between subtype A and C and between A and D have been reported from Eastern African countries which include Tanzania, Zambia, Uganda and Kenya, where subtype A, C and D usually circulate6162. The prevalence of unique recombinant forms (URFs) and circulatory recombinant forms (CRFs) were also reported from different part of India30. URF B/C was reported from mainly north eastern part (Manipur) with different ancestral origin and breakpoint and B/C/D recombinants from Punjab, indicating that these URFs and CRFs may evolve through mixing of the co-existing strains or enter through trafficking due to the close geographical proximity. The AC recombinant was identified from Maharashtra in India, where subtype C and A co-circulate63. The prevalence of CRFs and URFs indicates the changing patterns of HIV-1 epidemiology in India. The nature of HIV-1 circulating in different parts of India has been shown so far in the context of specific HIV-1 structural genes env and gag64–66. A report from north India showed the prevalence of multiple subtype including B’ (Thai B), C and A which indicates that the genetic diversity of HIV in India is gradually expanding67. Since HIV-1 recombination occurs in a random manner, it is increasingly likely that cross-over breakpoints could also occur within the ORFs of the HIV-1 genes. Extensive intra-subtype recombination in South African HIV-1 subtype C genome68, supports the above possibility. The appearance of CRF_02AG envelope sequences was reported for the first time by our group. This observation is important because majority of the earlier studies suggest that subtype C isolates originated from Southern Africa whereas CRF_02AG is predominantly found in Central and West Africa. Three independent reports from Indian investigators suggest that it is possible to occasionally find an X4-tropic virus among subtype C infected individuals and this happens with much reduced frequency as compared to subtype B which exceeds 50 per cent34–36. Earlier V3 to V5 regions of subtype C was reported to contribute to higher levels of HIV-1 replication69. Also co-infections (mycobaterial and viral) among HIV-1 infected individuals in India is rampant and may significantly alter the course of HIV-1 progression. In this connection, our group for the first time showed that the X gene of hepatitis B virus could augment HIV-1 LTR C promoter driven expression significantly more than subtype B LTR promoter and that Tat + HBx gene could act synergistically to activate HIV-1 gene expression70. Our observation that Vpu C causes more apoptosis than Vpu B may help explain some of the features associated with subtype C specific pathogenesis.
The appearance of new recombinants from several regions of India has potential to impact on the largely subtype-C driven epidemic. How they eventually impact on the overall fitness of the virus will be important for several reasons including possibility of generating new epitopes important for eliciting protective immune responses.
The genetic information of accessory genes from India also reveals the presence of multiple subtypes. All earlier studies with respect to recombinant viruses from India suggested any one of the ten viral genes to be related to a different subtype, for e.g. HIV-1 gag could be related to subtype C but the envelope from subtype B. Our studies clearly show that multiple cross-over events can occur within the open reading frame of a single HIV-1 gene.
The work by our group at National Institute of Immunology, New Delhi, was supported by Department of Biotechnology, Indian Council of Medical Research (ICMR), National Bioscience Award of Department of Biotechnology, Government of India to the last author (ACB) and via ICMR-NIH-RO3 grant. Generous help with several HIV-1 research related materials from AIDS Research & Reference Reagent Program, NIH, MD, USA, is gratefully acknowledged.
References
- Evidence for HTLV-III infection in prostitutes in Tamil Nadu (India) Indian J Med Res. 1987;85:335-8.
- [Google Scholar]
- NACO (home page on the internet). HIV Sentinel surveillance and HIV estimation in India. 2007. Available from: http:// www.nacoonline.org/upload/Publication/M&E%20Surveillance,%20Research/HIV%20Sentinel%20Surveillance%20and%20HIV%20Estimation%202007_A%20Technical%20Brief.pdf
- [Google Scholar]
- HIV infection in India: epidemiology, molecular epidemiology and pathogenesis. J Biosci. 2008;33:515-25.
- [Google Scholar]
- The Indian pediatric HIV epidemic: a systematic review. Curr HIV Res. 2008;6:419-32.
- [Google Scholar]
- HIV/AIDS epidemic in India: risk factors, risk behaviour & strategies for prevention & control. Indian J Med Res. 2005;121:356-68.
- [Google Scholar]
- Global HIV-1 molecular epidemiology with special reference to genetic analysis of HIV-1 subtypes circulating in North India: functional and pathogenic implications of genetic variation. Indian J Exp Biol. 2009;47:424-31.
- [Google Scholar]
- Host genes that affect progression of AIDS/HIV in India and novel gene therapeutic approaches against HIV. Indian J Biochem Biophys. 2008;45:141-8.
- [Google Scholar]
- Timing and reconstruction of the most recent common ancestor of the subtype C clade of human immunodeficiency virus type 1. J Virol. 2004;78:10501-6.
- [Google Scholar]
- Genomic diversity of human immunodeficiency virus type-1 in India. Int J STD AIDS. 2002;13:115-8.
- [Google Scholar]
- HIV type 1 subtypes circulating in eastern and northeastern regions of India. AIDS Res Hum Retroviruses. 2002;18:1219-27.
- [Google Scholar]
- Full-length gag sequences of HIV type 1 subtype C recent seroconverters from Pune, India. AIDS Res Hum Retroviruses. 2004;20:1113-8.
- [Google Scholar]
- Transmission of genetically diverse strains of HIV-1 in Pune, India. Indian J Med Res. 1998;107:1-9.
- [Google Scholar]
- Genomic diversity in the regulatory nef gene sequences in Indian isolates of HIV type 1: emergence of a distinct subclade and predicted implications. AIDS Res Hum Retroviruses. 2006;22:1206-19.
- [Google Scholar]
- Identification of subtype C human immunodeficiency virus type 1 by subtype-specific PCR and its use in the characterization of viruses circulating in the southern parts of India. J Clin Microbiol. 2004;42:2742-51.
- [Google Scholar]
- A genetic analysis of HIV-1 from Punjab, India reveals the presence of multiple variants. AIDS. 1995;9:685-90.
- [Google Scholar]
- Detection of HIV type 1 env subtypes A, B, C, and E in Asia using dried blood spots: a new surveillance tool for molecular epidemiology. AIDS Res Hum Retroviruses. 1996;12:1435-41.
- [Google Scholar]
- Subtype B and subtype C HIV type 1 recombinants in the northeastern State of Manipur, India. AIDS Res.Hum.Retroviruses. 2005;21:152-7.
- [Google Scholar]
- Genetic and functional characterization of the LTR of HIV-1 subtypes A and C circulating in India. AIDS Res.Hum.Retroviruses. 2007;23:1428-33.
- [Google Scholar]
- Phylogenetic analysis of HIV-1 reverse transcriptase sequences from 382 patients recruited in JJ Hospital of Mumbai, India, between 2002 and 2008. AIDS Res Hum Retroviruses. 2009;25:633-5.
- [Google Scholar]
- Novel HIV-1 long terminal repeat (LTR) sequences of Subtype B and mosaic intersubtype B/C recombinants in North India. Arch Virol. 2008;153:1961-6.
- [Google Scholar]
- Alterations in HIV-1 LTR promoter activity during AIDS progression. Virology. 2003;317:109-18.
- [Google Scholar]
- Multifacted activities of the HIV-1 transactivator of transcription, Tat. J Biol Chem. 1999;274:28837-40.
- [Google Scholar]
- Tat protein of human immunodeficiency virus type 1 subtype C strains is a defective chemokine. J Virol. 2004;78:2586-90.
- [Google Scholar]
- Clade-specific differences in neurotoxicity of human immunodeficiency virus-1 B and C Tat of human neurons: significance of dicysteine C30C31 motif. Ann.Neurol. 2008;63:366-76.
- [Google Scholar]
- Functional analysis of HIV-1 subtypes B and C HIV-1 Tat exons and RGD/QGD motifs with respect to Tat-mediated transactivation and apoptosis. AIDS. 2008;22:1683-5.
- [Google Scholar]
- Phase I therapeutic trial of the HIV-1 Tat protein and long term follow-up. Vaccine. 2009;27:3306-12.
- [Google Scholar]
- Isolation of a human gene that inhibits HIV-1 infection and is suppressed by the viral Vif protein. Nature. 2002;418:646-50.
- [Google Scholar]
- Full-length human immunodeficiency virus type 1 genomes from subtype C-infected seroconverters in India, with evidence of intersubtype recombination. J Virol. 1999;73:152-60.
- [Google Scholar]
- C-terminal half of HIV-1 Vif C possesses major determinant for APOBEC3G degradation. AIDS. 2009;23:141-3.
- [Google Scholar]
- The Vpr protein from HIV-1: distinct roles along the viral life cycle. Retrovirology. 2005;2:11.
- [Google Scholar]
- Genetic and functional characterization of human immunodeficiency virus type-1 VprC variants from north India: Presence of unique recombinants with mosaic genomes from B, C and D subtypes within the open reading frame of Vpr. J Gen Virol. 2009;90:2768-76.
- [Google Scholar]
- Role of HIV-1 subtype C envelope V3 to V5 regions in viral entry, coreceptor utilization and replication efficiency in primary T-lymphocytes and monocyte-derived macrophages. Virol J. 2007;4:126.
- [Google Scholar]
- Exceptional molecular and coreceptor-requirement properties of molecular clones isolated from an human immunodeficiency virus type-1 subtype C infection. Retrovirology. 2008;5:25.
- [Google Scholar]
- HIV-1 clade C env clones obtained from an Indian patient exhibiting expanded coreceptor tropism are presented with naturally occurring unusual amino acid substitutions in V3 loop. Virus Res. 2009;144:306-14.
- [Google Scholar]
- Genetic analysis of HIV-1 circulating recombinant form 02_AG, B and C subtype-specific envelope sequences from Northern India and their predicted co-receptor usage. AIDS Res Ther. 2009;6:28.
- [Google Scholar]
- Evolution of CCR5 use before and during coreceptor switching. J Virol. 2008;82:11758-66.
- [Google Scholar]
- First report of a healthy Indian heterozygous for delta32 mutant of HIV-1 coreceptor-CCR5 gene. Gene. 1998;207:141-7.
- [Google Scholar]
- Sequence specific cleavage of the HIV-1 coreceptor CCR5 gene by a hammer-head ribozyme and a DNA-enzyme: inhibition of the coreceptor function by DNA-enzyme. FEBS Lett. 1998;436:233-8.
- [Google Scholar]
- Genetic and functional analysis of HIV-1 Rev Responsive Element (RRE) sequences from North India. AIDS Res Ther. 2010;7:29.
- [Google Scholar]
- Inhibition of β-TrcP-dependent ubiquitination of p53 by HIV-1 Vpu promotes p53 mediated apoptosis in human T cells. Blood. 2011;117:6600-7.
- [Google Scholar]
- Retrovirus mutation rates and their role in genetic variation. J Gen Virol. 1998;79:1337-45.
- [Google Scholar]
- Recombinant HIV sequences: Their role in the global epidemic. In: Kuiken C, Foley B, Hahn B, Korber B, McCutchan F, Marx P, eds. Human retroviruses and AIDS: a compilation and analysis of nucleic acid and amino acid sequences. Los Alamos, New Mexico: Los Alamos National Laboratory; 2000. p. :54-72.
- [Google Scholar]
- Global molecular epidemiology of HIV: Understanding the genesis of AIDS pandemic. Adv Pharmacol. 2008;56:1-25.
- [Google Scholar]
- Identification and characterization of a new class of human immunodeficiency virus type 1 recombinants comprised of two circulating recombinant forms, CRF07_BC and CRF08_BC, in China. J Virol. 2003;77:685-95.
- [Google Scholar]
- Structure-functional analysis of human immunodeficiency virus type 1 (HIV-1) Vpr: role of leucine residues on Vpr-mediated transactivation and virus replication. Virology. 2004;328:89-100.
- [Google Scholar]
- The distribution of HIV-1 recombination breakpoints. Infect Genet Evol. 2007;7:717-23.
- [Google Scholar]
- Molecular mechanisms of recombination restriction in the envelope gene of the human immunodeficiency virus. PLoS Pathog. 2009;5(5):e1000418.
- [Google Scholar]
- A genetic-algorithm approach to simulating human immunodeficiency virus evolution reveals the strong impact of multiply infected cells and recombination. J Gen Virol. 2005;86:3109-18.
- [Google Scholar]
- Extensive recombination among human immunodeficiency virus type 1 quasispecies makes an important contribution to viral diversity in individual patients. J Virol. 2006;80:2472-82.
- [Google Scholar]
- Stochastic interplay between mutation and recombination during the acquisition of drug resistance mutations in human immunodeficiency virus type 1. J Virol. 2005;79:13572-8.
- [Google Scholar]
- Recombination leads to the rapid emergence of HIV-1 dually resistant mutants under selective drug pressure. Proc Natl Acad Sci. U S A. 1996;93:6106-111.
- [Google Scholar]
- Contribution of recombination to the evolution of human immunodeficiency viruses expressing resistance to antiretroviral treatment. J Virol. 2007;81:7620-8.
- [Google Scholar]
- Recombination increases human immunodeficiency virus fitness, but not necessarily diversity. J Gen Virol. 2008;89:1467-77.
- [Google Scholar]
- Dynamics of HIV-1 recombination in its natural target cells. Proc Natl Acad Sci USA. 2004;101:4204-9.
- [Google Scholar]
- Emergence of recombinant forms of HIV: dynamics and scaling. PLoS Comput Biol. 2007;3:2003-18.
- [Google Scholar]
- Mechanisms of nonrandom human immunodeficiency virus type 1 infection and double infection: preference in virus entry is important but is not the sole factor. J Virol. 2005;79:4140-9.
- [Google Scholar]
- High rates of human immunodeficiency virus type 1 recombination: near-random segregation of markers one kilobase apart in one round of viral replication. J Virol. 2003;77:11193-200.
- [Google Scholar]
- Evolution and probable transmission of intersubtype recombinant human immunodeficiency virus type 1 in a Zambian couple. J Virol. 1997;71:2647-55.
- [Google Scholar]
- Molecular characterization of HIV type 1 isolates from untreated patients of Mumbai (Bombay), India, and detection of rare resistance mutations. AIDS Res Hum Retroviruses. 2004;20:1032-5.
- [Google Scholar]
- Human immunodeficiency virus type 1 env sequences from Calcutta in eastern India: identification of features that distinguish subtype C sequences in India from other subtype C sequences. J Virol. 2002;75:10479-87.
- [Google Scholar]
- Phylogenetic analysis of the p24–p7 region of the human immunodeficiency virus type 1 gag gene to determine subtype distribution among female sex workers in Calcutta, India. J Clin Microbiol. 2005;43:5787-91.
- [Google Scholar]
- Polymorphism of HIV-1 gag (p17) gene from female sex workers in Calcutta, India. Arch Virol. 2005;150:2117-24.
- [Google Scholar]
- Subtypes of HIV type 1 circulating in India: partial envelope sequences. AIDS Res Hum Retroviruses. 1999;15:941-4.
- [Google Scholar]
- Extensive intrasubtype recombination in South African human immunodeficiency virus type-1 subtype C infections. J Virol. 2007;81:4492-500.
- [Google Scholar]
- Role of HIV-1 subtype C envelope V3 to V5 regions in viral entry, coreceptor utilization and replication efficiency in primary T-lymphocytes and monocyte-derived macrophages. Virol J. 2007;4:126.
- [Google Scholar]
- X protein of hepatitis B virus potently activates HIV-1 subtype C long terminal repeat promoter: implications for faster spread of HIV-1 subtype C. AIDS. 2007;21:1491-2.
- [Google Scholar]






