Translate this page into:
Emergence of vanA gene among vancomycin-resistant enterococci in a tertiary care hospital of North - East India
Reprint requests: Dr Chimanjita Phukan, Department of Microbiology, Gauhati Medical College & Hospital, Guwahati 781 005, Assam, India e-mail: chimanjitaphukan@gmail.com
-
Received: ,
This is an open access article distributed under the terms of the Creative Commons Attribution NonCommercial ShareAlike 3.0 License, which allows others to remix, tweak, and build upon the work non commercially, as long as the author is credited and the new creations are licensed under the identical terms.
This article was originally published by Medknow Publications & Media Pvt Ltd and was migrated to Scientific Scholar after the change of Publisher.
Abstract
Background & objectives:
Vancomycin-resistant enterococci (VRE) have become one of the most challenging nosocomial pathogens with the rapid spread of the multi-drug resistant strain with limited therapeutic options. It is a matter of concern due to its ability to transfer vancomycin resistant gene to other organisms. The present study was undertaken to determine the emergence of vancomycin-resistant enterococci and the vanA gene among the isolates in a tertiary care hospital of North-East India.
Methods:
A total of 67 consecutive enterococcal isolates from different clinical samples were collected and identified by using the standard methods. Antibiogram was done by disk diffusion method and VRE was screened by the disk diffusion and vancomycin supplement agar dilution method. The minimum inhibitory concentration (MIC) value for vancomycin was determined by E-test. The VRE isolates were analyzed by PCR for vanA gene.
Results:
A total of 54 (81%) Enterococcus faecalis and 13 (19%) E. faecium were detected among the clinical isolates and 16 (24%) were VRE. The VRE isolates were multidrug resistant and linezolid resistance was also found to be in three. MIC range to vancomycin was 16-32 µg/ml among the VRE. The vanA gene was found in nine of 16 VRE isolates.
Interpretation & conclusions:
Emergence of VRE and presence of vanA in a tertiary care hospital setting in North-East India indicate toward a need for implementing infection control policies and active surveillance.
Keywords
Enterococcus faecalis
Enterococcus faecium
nosocomial infection
vancomycin-resistant enterococci (VRE)
VanA gene
Enterococci have emerged as the leading causes of multiple drug resistant hospital-acquired pathogens especially with the emergence of glycopeptide-resistant enterococcus (GRE) species. It is the second most common pathogen causing mortality and morbidity1 and the third leading cause of hospital acquired bloodstream infection2. These not only pose challenge to the clinicians but also result in treatment failure, selection pressure and spread of resistant strains in the health care institutes.
GRE strains were observed after almost 25 years of vancomycin use since 1980s. In the United States, vancomycin-resistance Enterococcus faecium accounted for 4 per cent of healthcare-associated infections3. In Europe, prevalences of VRE is diverse ranging from <1 to >40 per cent4. Resistance to glycopeptides is mediated by the van gene clusters, which produce resistance by altering the drug target from D-alanine-D-alanine to D-alanine-D-lactate5. So far, eight genotypes of glycopeptide resistance, which differ in the level and range of resistance and in transferability to glycopeptides, have been described for enterococci. Five of the van genes are acquired (vanA, B, D, E, G) and three (vanC1, C2, C3) are intrinsic. Of these, vanA is the most prevalent and is predominantly found in E. faecium and E. faecalis, the enterococcal species responsible for most infections in human6. This study was undertaken to detect the presence of vancomycin resistant enterococci (VRE) and that of vanA among the enterococcal isolates in a tertiary care hospital in North-East (NE) India.
Material & Methods
A total of 67 consecutive, non-repetitive clinical isolates of Enterococcus species from different clinical samples (urine, blood, sputum, pus and throat swabs) were obtained in the department of Microbiology and this study was carried out in the Genetic Laboratory, under department of Microbiology, Gauhati Medical College and Hospital and National Institute of Pharmaceutical and Education and Research, Guwahati. The approval from the institutional ethical committee was obtained prior to the study.
Enterococcus species were identified in accordance with the standard procedures5.
The isolates were stored for further processing in Luria Bertani (LB) broth with 20 per cent glycerol at -80°C.
Antibiotic susceptibility testing: Antibiotic susceptibility testing was done using Kirby-Bauer disk diffusion method as per the CLSI (Clinical Laboratory Standards Institute) guideline7 against vancomycin(30 μg), teicoplanin (30 μg), linezolid (30 μg), penicillin G (10 U), tetracycline (30 μg), ciprofloxacin (30 μg), chloramphenicol (30 μg) and doxycycline (30 μg). The quality control organisms were E. faecalis ATCC 51299 (VRE) and E. faecalis ATCC 29212. (Hi-media, Mumbai).
Phenotypic detection of vancomycin resistant Enterococcus: Vancomycin resistance in all enterococcal isolates was detected by disk diffusion method and VRE agar dilution screening test7. Vancomycin supplement (6 μg, Hi-media, Mumbai) was added to 1 ml of sterile distilled water and then mixed to 1000 ml of the autoclaved brain heart infusion (BHI) agar at 45°C and the medium was pour in a petridish and the bacteria was inoculated and incubated at 37°C for 48 h. Growth of even single colony was considered as vancomycin-resistant Enterococcus (VRE).
MIC for vancomycin-resistant Enterococcus: Minimum inhibitory concentration (MIC) value for vancomycin was determined using E-test strip (Hi-media, Mumbai). Any Enterococcus was considered VRE if the MIC was ≥ 16 μg/ml7.
Polymerase chain reaction (PCR) for vanA gene: The isolates resistant to vancomycin were taken for plasmid DNA isolation and amplification. A single colony was picked from a freshly streaked blood agar plate and inoculated in 3 ml L-B broth where it was grown at 37°C for 12-16 h with constant shaking. The culture was incubated for 1 h at 37°C with lysozyme (5 mg/ml). The bacterial cells were harvested by centrifugation at 6000 x g for 15 min at 4°C. The pellet was taken and resuspendend in tris-acetate-EDTA (TAE) buffer and heated in a heat block at 95°C for 5 min. It was further centrifuged at in ‘g’ 15000×g for 5 min at 4°C and the supernatant containing DNA was used as a source of template for amplification.
The PCR amplification for vanA was performed8 with some modifications. The reaction mixture with a final volume of 50 μl contained 3 μl of purified plasmid DNA, 1 × PCR buffer (20 mM Tris-HCl/50 mM KCl, pH 8.4), 7 mM MgCl2, 0.2 mM each deoxynucleoside triphosphate, 0.5 nM each primer, and 2.5 units of Taq polymerase (Qiagen, Mumbai). The PCR conditions were 95° C for 5 min for the first cycle; 95°C for 1 min, 50°C for 1 min and 720C for 1 min for the next 30 cycles and final extension at 72°C for 10 min.
The vanA gene primer sequence (5’-3’) used was: forward primer - A1 GGGAAAACGACAA TTGC and reverse primer -A2 GTACAATGCGGCCGTTA9. Amplification of gene was carried out by DNA thermal cycler with the help of specific primers for vanA gene. E. faecalis ATCC 51299 was used as positive control for vanA and the negative control consisted of all reagents but no DNA template. PCR product was analyzed by electrophoresis with 1.5 per cent agarose gel and 0.5 mg ethidium bromide added separately. Gel was photographed on an ultraviolet light transilluminator by gel documentation system (Perkin Elmer, USA). 100 bp molecular marker was used and a final product of 732 bp was considered as vanA gene.
Results & Discussion
A total of 67 enterococcal isolates were collected from 896 culture-positive samples screened over the one year period. The majority of isolates were recovered from urine specimens followed by isolates from blood, sputum, pus and throat swab; 49 (73%) of the enterococcal isolates were obtained from hospitalized patients and 18 (27%) were from outpatient department. Among these 67 enterococcal isolates, 16 (24%) VRE and 51 (76%) vancomycin sensitive enterococcus (VSE) were detected (Table I); 54 (81%) were E. faecalis and 13 (19%) E. faecium and among the VRE, nine (56.3%) E. faecalis and seven (43.7%) E. faecium were detected.
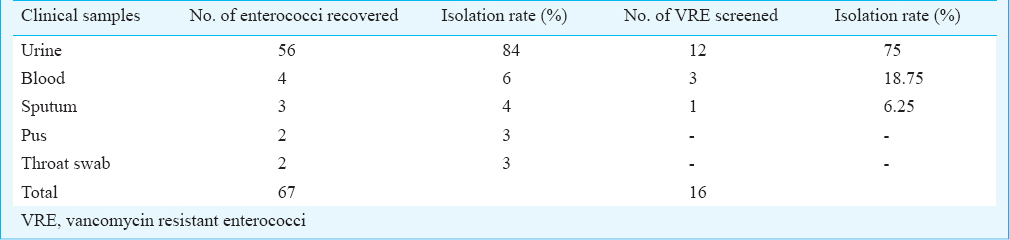
Of the 67 isolates, 46 (68.6%) were multiple drug resistant to more than one class of antibiotics, three (4.5%) were resistant to linezolid (Table II) and were found to be VRE. The linezolid resistance was found in two E. faecalis and one E. faecium VRE isolates. The VRE isolates were resistant to ciprofloxacin (69%), tetracycline (56%), penicillin (58%) and erythromycin (100%). Most of the VRE were found to be sensitive to tecoplanin and linezolid (Fig. 1). Among the VRE, MIC to vancomycin ranged between 16-32 µg/ml.

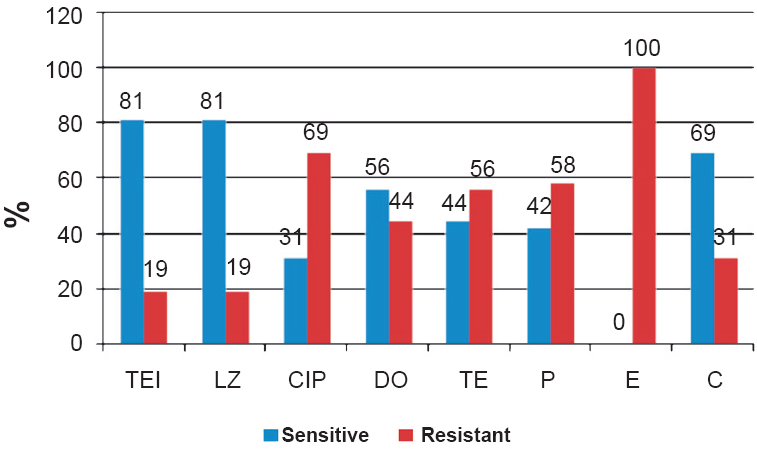
- Antibiogram sensitivity of VRE isolates. TEI, teicoplanin; LZ, linezolid; CIP, ciprofloxacin; DO, doxycycline; TE, tetracycline; P, penicillin; E, erythromycin; C, chloramphenicol.
Nine of 16 VRE isolates were found to possess VanA gene with a 732 bp PCR product (Fig. 2) of which six were E. faecium and three E. faecalis.
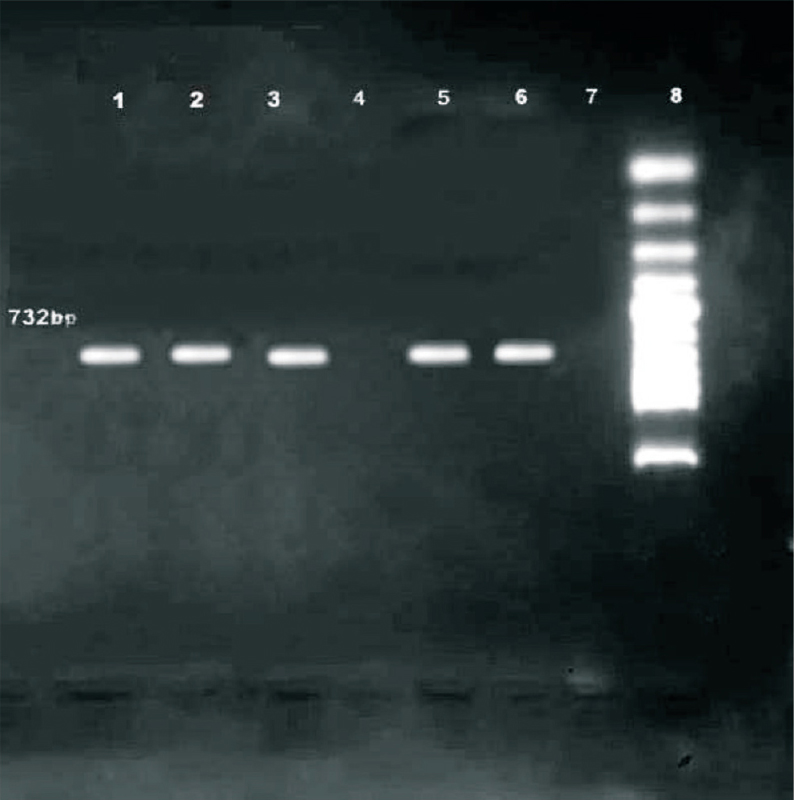
- Agarose gel electrophoresis showing amplification of 723 bp fragment for vanA of VRE from the clinical samples. Lanes 1-3, 5-6: VRE strains represents the vanA amplified 732 bp fragment, Lane 7 - E. faecium ATCC 29212 (negative control), Lane 8-100 bp ladder marker.
In the present study, 81 per cent E. faecalis and 19 per cent E. faecium were detected. Earlier studies from various parts of India had shown 55-87 per cent of E. faecalis as the predominant species followed by 10-20 per cent of E. faecium10111213. In some studies a relatively high proportion of E. faecium has been reported1415. In our study maximum isolation was from the urine followed by blood. This finding was consistent with other studies121617.
A study from Lucknow reported VRE in 55.17 per cent of the isolates18. Other Indian studies reported VRE isolation between 0-5 per cent101113. Studies from Indore and Nagpur reported 14.29 and 11.38 per cent VRE, respectively1920. In our study the VRE isolation was 24 per cent.
Studies from New Delhi reported MIC range of vancomycin >32 μg/ml10, from Chandigarh 8-16 μg/ml11 and from Mumbai 8-128 µg/ml13. Studies from Italy21, Japan22 and Turkey23 showed MIC range of vancomycin 16-32 μg/ml similar to our study.
The vanA gene was found in 56.25 per cent of the VRE isolates in our study. Mathur et al10 reported 80 per cent of vanA phenotype which was higher than our findings. Studies from Taiwan24 and Brazil25 reported vanA gene in all the VRE isolates. The steady rise in the proportion of vancomycin-resistant E. faecalis isolates in the United States from 2.7 per cent in 1999 to 3.9 per cent in 2010 has been reported26.
In conclusion, emergence of VRE and vanA gene among the isolates in a tertiary care hospital setting in NE India points towards implementing prevention and control policies.
References
- Prevalence and antimicrobial resistance pattern of multidrug- resistant enterococci isolates from clinical specimens. Indian J Med Microbiol. 2012;30:44-51.
- [Google Scholar]
- Emergence of Enterococcus as a significant pathogen. Clin Infect Dis. 1992;14:1173-8.
- [Google Scholar]
- NHSN annual update: antimicrobial-resistant pathogens associated with healthcare- associated infection: annual summary of data reported to the national healthcare safety network at centers for disease control and prevention 2006-2007. Infect Control Hosp Epidemiol. 2008;29:996-1011.
- [Google Scholar]
- Emergence and spread of vancomycin resistance among enterococci in Europe. Euro Surveill. 2008;13((47)) pii=19046
- [Google Scholar]
- Vancomycin resistance in gram-positive cocci. Clin Infect Dis. 2006;42((Suppl 1)):S25-34.
- [Google Scholar]
- Clinical Laboratory Standards Institute (CLSI). Performance standards for antimicrobial susceptibility testing, 21st informational supplement (M100-S21) Wayne, PA: CLSI; 2011.
- [Google Scholar]
- Detection of glycopeptide resistance genotypes and identification to the species level of clinically relevant enterococci by PCR. J Clin Microbiol. 1995;33:24-7.
- [Google Scholar]
- The VanA glycopeptide resistance protein is related to D-alanyl-D-alanine ligase cell wall biosynthesis enzymes. Mol Gen Genet. 1990;224:364-72.
- [Google Scholar]
- Antimicrobial resistance in Enterococcus fecalis at a tertiary care center of Northern India. Indian J Med Res. 2003;118:25-8.
- [Google Scholar]
- Significance of the vancomycin resistant enterococci which were isolated from urinary specimens at a tertiary care centre in north India. Indian J Med Res. 2004;119:72-4.
- [Google Scholar]
- Enterococcal infections with special reference to the phenotypic characterization and the drug resistance. Indian J Med Res. 2004;119:22-5.
- [Google Scholar]
- Vancomycin resistant enterococci in a tertiary care hospital in Mumbai. Indian J Med Microbiol. 2009;27:375-6.
- [Google Scholar]
- Change in the prevalence and the antibiotic resistance of the enterococcal species isolated from blood cultures. J Clin Diag. 2012;6:405-8.
- [Google Scholar]
- Clinico-epidemiological profile and high-level aminoglycoside resistance in enterococcal septicemia from a tertiary care hospital in east Delhi. Int J App Basic Med Res. 2011;1:80-3.
- [Google Scholar]
- Molecular screening of virulence genes in high-level gentamicin-resistant Enterococcus faecalis and Enterococcus faecium isolated from clinical specimens in Northwest Iran. Indian J Med Microbiol. 2012;30:175-81.
- [Google Scholar]
- Antibiotic options for Enterococcus faecalis infections. Pak J Med Sci. 2006;22:286-90.
- [Google Scholar]
- A new approach of real time polymerase chain reaction in detection of vancomycin-resistant enterococci and its comparison with other methods. Indian J Med Microbiol. 2013;31:47-52.
- [Google Scholar]
- In vitro activity of daptomycin & linezolid against methicillin resistant Staphylococcus aureus & vancomycin-resistant enterococci isolated from hospitalized cases in Central India. Indian J Med Res. 2013;137:191-6.
- [Google Scholar]
- Study of antimicrobial resistance in enterococci. Indian J Med Microbiol. 2008;26:285-7.
- [Google Scholar]
- Use of primers selective for vancomycin resistance genes to determine van genotype in enterococci and to study gene organization in VanA isolates. Antimicrob Agents Chemother. 1995;39:1772-8.
- [Google Scholar]
- Rapid detection and differentiation method of VanA, VanB and VanC phenotypes in vancomycin-resistant enterococci. Int J Antimicrob Agents. 2004;23:502-5.
- [Google Scholar]
- Molecular characterization of antibiotic resistant Enterococci. Int J Infect Dis. 2008;12:e273.
- [Google Scholar]
- Emergence of vancomycin-resistant Enterococci at a University Hospital in Taiwan: Persistence of multiple species and multiple clones. Infect Control Hosp Epidemiol. 1999;20:828-33.
- [Google Scholar]
- Molecular typing and antimicrobial susceptibility of vancomycin-resistant Enterococcus faecium in Brazil. Infect Control Hosp Epidemiol. 2002;2:19-22.
- [Google Scholar]
- East north central region has the highest prevalence of vancomycin-resistant Enterococcus faecalis in the United States. Infect Control Hosp Epidemiol. 2013;34:443-5.
- [Google Scholar]






