Translate this page into:
Emergence of multi drug resistant bacteria in diabetic patients with lower limb wounds
† For correspondence: gopic@aims.amrita.edu
This is an open-access article distributed under the terms of the Creative Commons Attribution-Noncommercial-Share Alike 3.0 Unported, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
This article was originally published by Medknow Publications & Media Pvt Ltd and was migrated to Scientific Scholar after the change of Publisher.
Sir,
Lower limb infections are the largest non-traumatic cause of lower extremity amputations in diabetic patients, accounting for almost 90,000 amputations per year12. Polymicrobial infections are associated with an increased risk of amputations, prolonged hospital stay, increased expenses and higher infection-related mortality3. Ramakant et al4 reported 66 per cent cultures as polymicrobial and 23 per cent monomicrobial in diabetic wounds. Predominant bacterial isolates were Enterococcus faecalis, Staphylococcus aureus, Pseudomonas aeruginosa and Escherichia coli45. Culture specific antimicrobial therapy coupled with aggressive surgical excision of the necrotic tissue remains the gold standard for managing infections6. Yet, the wound healing, recurrence of infection and lower extremity amputation rates are increasing remarkably among diabetic patients7. Widespread use of antibiotics along with the natural evolution of bacteria has led to a number of multi-drug resistant bacteria (MDRB)89. Acquisition of resistance has become one of the major causes of treatment failure. In contrast, none of the antibacterial agents used in clinical trials possessed sufficiently novel modes of action to circumvent extant antibiotic resistance mechanisms10.
The emergence of MDRBs in diabetic wounds is less studied in Indian population. We, therefore, enrolled 591 consecutive diabetic patients with lower limb wounds who were hospitalized from January 2012 to December 2012 at the Podiatry Center, Amrita Institute of Medical Sciences and Research Center, Kochi, India. Deep tissue was collected from the wound bed under sterile precaution in the operation theater; and bacterial culture was tested against 40 antibiotics (eg. Colistin, tigecyclin, teicoplanin, clindamycin, meropenem, amoxicillin ciprofloxacin) as per the Clinical and Laboratory Standards Institute (CLSI) guidelines11. Bacterial culture and sensitivity were not done for 47 patients as they were hospitalized for correcting their foot deformities (e.g. Charcot foot, Hallux valgus, Equinus deformity). Of the remaining 544 patients, 416 (76.5%) were males and 128 (23.5%) were females. The mean age of the patients was 61.24±10.38years, HbA1c:9.39±2.27%, platelet count:3.54±1.48×106/μl and neutrophils:68.1±24.3%. Bacterial infections were found in (448/544, 82.3%) patients and 96(17.7%) patients had no bacterial growth. Predominant bacteria isolated from wounds were E. coli (73/448, 16.3%), P. aeruginosa (48/448, 10.7%), E. faecalis (42/448, 9.2%), Klebsiella pneumoniae (40/448, 8.9%), S. aureus (40/448, 8.9%) and Coagulase negative Staphylococcus (36/448, 8.0%), Acinobacter baumannii (23/448, 5.1%), Enterobacter sp. (22/448, 4.9%), Proteus mirabilis (15/448, 3.3%), and Streptococcus sp. (14/448, 3.1%). Common surgical interventions done were wound debridement: (200/591, 34%), toe amputation: (118/591, 20%), below knee amputation: (60/591, 10.2%), mid-foot amputation: (18/59, 8.3%) and above knee amputations: (18/591, 2.9%) and other minor surgeries (146/591, 24%). E.coli, K. pneumoniae, E. faecalis, P. mirabilis and A. baumannii and S. auricularis were predominant bacterial isolates found in patients who underwent above or below knee amputations. The figure shows the per cent of MDRBs in diabetic lower limb wounds.
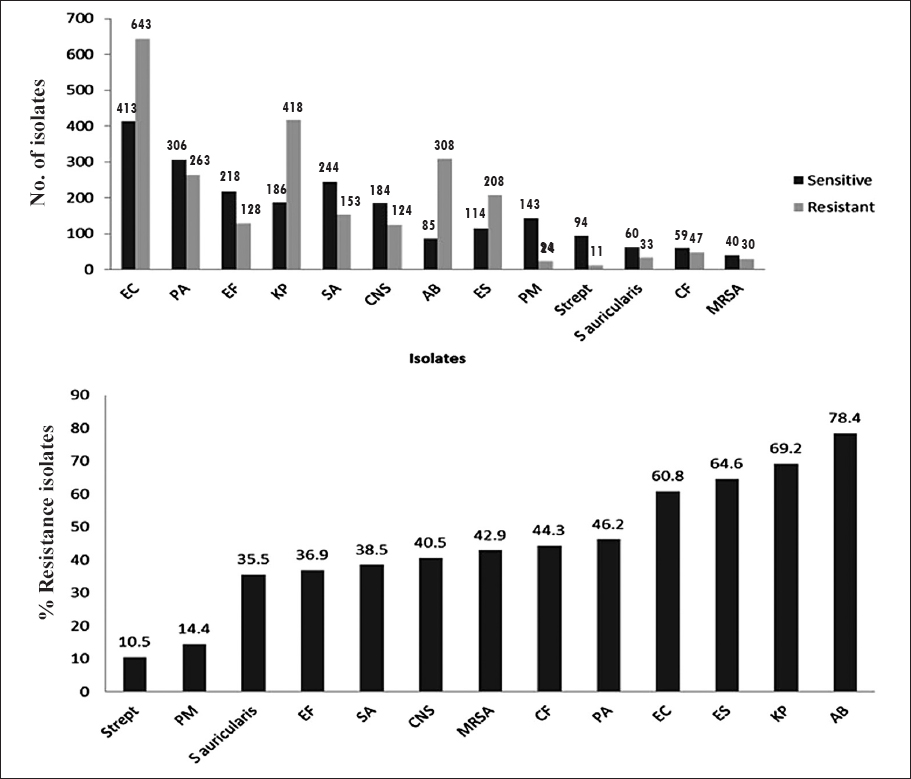
- Most predominant multi drug resistant bacteria (MDRBs) isolated from lower limb wound of patients with diabetes and their antimicrobial resistance. EC, Escherichia coli; PA, Pseudomonas aeruginosa; EF, Enterococcus faecalis; KP, Klebsiella pneumoniae; SA, Staphylococcus aureus; CNS, coagulase negative staphylococci; AB, Acinobacter baumanii; ES, Enterobacter sp; PM, Proteus mirabilis; Strept, Streptococcus sp; CF, Citrobacter freundii; mRSA, methicillin resistant Staphylococcus aureus.
E. coli was susceptible to tigecyclin and colistin; P.aeruginosa and K. pneumoniae to colistin; E. faecalis to vancomycin, tigecyclin and linezolid. E. coli was resistant to beta lactam group (BLG), fluoroquinolones (FQ) and macrolides. P. aeruginosa was resistant to FQ and macrolides; and sensitive to colistin and carbapenem. E. fecalis showed resistance to penicillins and macrolides and susceptibility to vancomycin, tigecyclin and linezolid. K. pneumoniae was resistant to beta lactam group, FQ; and susceptible to macrolides and tigecyclin. S. aureus was resistant to macrolides; and sensitive to penicillin, FQ, and co-trimoxazole.
The study protocol was approved by the ethics committee of the institute. About a decade earlier, only the MRSA (15-30%) and MDR P. aeruginosa (44%) had been reported to be predominant in diabetic wounds14. We observed an increase in many other MDRBs in diabetic patients with lower limb wounds. Goldstein et al11 showed that patients who had previously received oral antibiotics were more likely to have MRSA, enterococci, and P. aeruginosa and less likely to have Enterobacteriacea and anaerobes isolated from their wounds. With the current expansion of the reservoir of resistant organisms, obtaining reliable deep cultures can help focus antimicrobial therapy against the dominant pathogens12. It is reported that while initiating antimicrobial therapy, vascular status of the lower limb, depth and severity of infection should be considered. When the diabetic wound is ischaemic, the concentration of antibiotics at the wound site may be insufficient to act against the bacteria. Hence the microbe may become resistant to the specific antibiotics13. On the other hand, it is practically difficult to administer higher doses of antibiotics when liver and kidney functions are compromised. Drug and dose related adverse events are the other limiting factors.
The population genetics of pathogenic bacteria has been extensively studied to understand the spread of disease and the evolution of virulence and drug resistance. However, little attention has been paid to bacterial carriage populations, which inhabit hosts without producing disease. Perron et al14 have found that asymptomatic swine from livestock productions frequently carry populations of Salmonella enterica with a broad range of drug-resistant strains and genetic diversity greatly exceeding from that previously described.
Another major mechanism for bacterial resistance to antibiotics is through the acquisition of a plasmid coding for resistance-mediating proteins. Many of the bacteria become resistant to antibiotics through the process of lateral gene transfer, with the newly acquired genes encoding a variety of resistance-mediating proteins15. This plasmid-encoded resistance has been observed for virtually all classes of antibiotics and in a wide variety of Gram-positive and Gram-negative organisms; many antibiotics are no longer effective due to such plasmid-encoded resistance15. The systematic removal of these resistance-mediating plasmids from the bacteria would re-sensitize bacteria to standard antibiotics15. Intravenous human immunoglobulin therapy was reported to augment opsonic activity against various drug-resistant bacteria, and being tried for treating severe bacterial infections in immunocompromised patients with impaired serum opsonic capacity16.
To conclude, an emergence of MDRBs was observed in lower limb wounds of patients with diabetes. Bacteria isolated from wounds were resistant to most of the antibiotics except colistin, vancomycin, linezolid and tegecyclin. Further studies are warranted to understand the reasons for antimicrobial resistance; and for reducing the morbidity and mortality related to wound infections caused by MDRBs.
Acknowledgment
Authors acknowledge the Amrita Vishwa Vidya Peetham University, Kochi for granting permission to conduct the study at Amrita Institute of Medical Sciences and Research Center.
References
- Bacterial etiology of diabetic foot infections in South India. Eur J Intern Med. 2005;16:567-70.
- [Google Scholar]
- Ertapenem versus piperacillin/tazobactam for diabetic foot infections (SIDESTEP): prospective, randomised, controlled, double-blinded, multicentre trial. Lancet. 2005;366:1695-703.
- [Google Scholar]
- Co-therapy using lytic bacteriophage and linezolid: effective treatment in eliminating methicillin resistant Staphylococcus aureus (MRSA) from diabetic foot infections. PLoS One. 2013;8:e56022.
- [Google Scholar]
- Changing microbiological profile of pathogenic bacteria in diabetic foot infections: time for a rethink on which empirical therapy to choose? Diabetologia. 2011;54:58-64.
- [Google Scholar]
- Spectrum and prevalence of fungi infecting deep tissues of lower-limb wounds in patients with type 2 diabetes. J Clin Microbiol. 2010;48:2097-102.
- [Google Scholar]
- Empirical therapy for diabetic foot infections: are there clinical clues to guide antibiotic selection? Clin Microbiol Infect. 2007;13:351-3.
- [Google Scholar]
- Diabetic foot infections: microbiology made modern? Array of hope. Diabetes Care. 2007;30:2171-2.
- [Google Scholar]
- Treating foot infections in diabetic patients: a randomized, multicenter, open-label trial of linezolid versus ampicillin-sulbactam/amoxicillin-clavulanate. Clin Infect Dis. 2004;38:17-24.
- [Google Scholar]
- Clinico-microbiological study and antimicrobial drug resistance profile of diabetic foot infections in North India. Foot (Edinb). 2011;21:6-14.
- [Google Scholar]
- Diabetic foot infections. Bacteriology and activity of 10 oral antimicrobial agents against bacteria isolated from consecutive cases. Diabetes Care. 1996;19:638-41.
- [Google Scholar]
- Management of diabetic foot infections in an era of increasing microbial resistance. Curr Infect Dis Rep. 2009;11:375-82.
- [Google Scholar]
- The combination of meropenem and levofloxacin is synergistic with respect to both Pseudomonas aeruginosa kill rate and resistance suppression. Antimicrob Agent Chemother. 2010;54:2646-54.
- [Google Scholar]
- A reservoir of drug-resistant pathogenic bacteria in asymptomatic hosts. PLoS One. 2008;3:e3749.
- [Google Scholar]
- Bacterial death comes full circle: targeting plasmid replication in drug-resistant bacteria. Org Biomol Chem. 2005;3:959-66.
- [Google Scholar]
- Opsonic activity assessment of human intravenous immunoglobulin preparations against drug-resistant bacteria. J Infect Chemother. 2004;10:234-8.
- [Google Scholar]





