Translate this page into:
Development & evaluation of biotinylated DNA probe for clinical diagnosis of chikungunya infection in patients’ acute phase serum & CSF samples
Reprint requests: Dr M.M. Parida, Division of Virology, Defence Research & Development Establishment, Jhansi Road, Gwalior 474 002, India e-amil: paridamm@rediffmail.com
-
Received: ,
This is an open-access article distributed under the terms of the Creative Commons Attribution-Noncommercial-Share Alike 3.0 Unported, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
This article was originally published by Medknow Publications & Media Pvt Ltd and was migrated to Scientific Scholar after the change of Publisher.
Abstract
Background & objectives:
The resurgence of chikungunya virus (CHIKV) in the Indian Ocean Islands and India has drawn worldwide attention due to its explosive nature, high morbidity and complex clinico-pathological manifestations. The early confirmatory diagnosis of CHIKV is essential for management as well as control of unprecedented epidemics. The present study describes the development and evaluation of a highly sensitive and specific E1 structural gene specific biotinylated DNA probe for detection of chikungunya virus in clinical samples using a dot blot format.
Methods:
The complementary DNA (cDNA) of CHIKV was spotted on to nylon membrane. The membrane was subjected to prehybridization and hybridization and developed using a colour development solution containing DAB chromogen.
Results:
The CHIKV E1 specific DNA probe was highly sensitive detecting picogram levels of target nucleic acid. The comparative evaluation with SYBR Green I based real-time RT-PCR revealed 99 per cent accordance with a sensitivity and specificity of 99 and 98 per cent, respectively. The specificity of this assay was further confirmed through cross-reaction studies with confirmed dengue and Japanese encephalitis (JE) patient serum samples along with infected culture supernatant of Ross River and Saint Louis encephalitis and plasmid DNA of O’Nyong Nyong, Semlinki forest and Sindbis viruses.
Interpretation & conclusion:
The DNA probe reported in this study may be useful for specific, sensitive and confirmatory clinical diagnosis of chikungunya infection in acute phase human patient serum and CSF samples. This assay can also be used in the laboratory for quantification of viral antigen in cell culture supernatant for research purpose.
Keywords
Biotinylated DNA probe
chikungunya
clinical diagnosis
E1 structural gene
Chikungunya fever is an acute arthropod borne viral illness reported in many parts of Africa and South East Asia1. The causative agent is chikungunya virus (CHIK V), a member of the genus Alphavirus of the family Togaviridae and is primarily transmitted by the Aedes aegypti and Ae. albopictus mosquitoes2. Chikungunya virus was first isolated from Tanzania in 19533, and later the virus has widely disseminated throughout sub-Saharan Africa, India, and countries of Southeast Asia, leading to numerous epidemics in the subsequent years4.
The genome of chikungunya virus consists of a linear, single stranded, positive sense ribonucleic acid (RNA) of approximately 11.7 kb in length. The Alphavirus genus consists of 30 species of arthropod borne viruses that can be classified antigenically into seven complexes (such as Barmah Forest, Eastern Equine Encephalitis, Middelburg, Ndumu, Semliki Forest,Venezuelan Equine Encephalitis and Western Equine Encephalitis) based on the envelope glycoprotein E1 and E2. The E2 protein is the site of neutralizing epitopes while the E1 protein contains more conserved cross-reactive epitopes5. Phylogenetic analysis on E1 gene sequences grouped CHIK viruses isolated worldwide into three genotypes: Asian, East/Central/South African (ECSA), and West African. In Africa, two genotypes viz. West African and East Central South African (ECSA) genotypes contributed to all the epidemics while the Asian genotypes represented for whole of Asia6. However, the 2005-2006 epidemics have seen the introduction of the ECSA genotype to the Asian continent for the first time. In Indian subcontinent, the outbreak has severely affected the areas of coastal States like Andhra Pradesh (AP), Kerala, Karanataka, Tamil Nadu and other States of Central India in 200678, and spread to 15 other States including Kerala. The complete genome sequence from the Kerala isolates revealed immense potential to cause severe infections as there was a shift from A226V in the E1 gene of the CHIKV9.
At present, there is no licensed vaccine or antiviral therapy available against chikungunya virus infection. Although not listed as a haemorrhagic fever virus, illness caused by CHIKV can be confused with diseases such as dengue or yellow fever based on the similarity of the symptoms. Thus, the differential diagnosis of these infections is essential for clinical management and epidemiological study in the tropics.
The routine laboratory diagnosis of CHIKV infection is based on culture, serology and also by identification of viral genome through reverse transcription polymerase chain reaction (RT-PCR)1011. However, confirmation of virus is based on 50 per cent reduction in plaque forming units using specific neutralizing antibody titre by plaque reduction neutralization (PRNT) assays12. Both virus isolation and PRNT assays are time consuming and tedious, requiring more than a week for completion. In addition, the IgM antibody that indicates recent infection usually appears by day 5 and thus is not useful for early clinical diagnosis13. Unlike dengue and Japanese encephalitis (JE) virus where low level and transient vireamia occurs, chikungunya virus produced high level of vireamia which persists up to 21 days of infection. Therefore, RT-PCR is the method of choice for early detection of CHIKV replication in supernatants and clinical samples14. In addition to conventional RT-PCR, there are more specific, sensitive and real-time PCR based assays such as TaqMan RT-PCR and loop mediated isothermal amplification (LAMP) methods have been reported which are currently under extensive evaluation with clinical samples1516.
Despite the high magnitude of amplification, these PCR based methods require either high precision instruments for the amplification or elaborate methods for detection of the amplified products. In addition, these methods are often cumbersome to adapt for routine clinical use especially in peripheral health care settings and private clinics. Considering the limitations of commercially available antibody detection systems and molecular methods such as RT-PCR for clinical diagnosis of chikungunya infection, a nucleic acid DNA probe offers the possibility of a sensitive, specific and reliable confirmatory diagnosis of virus infection at early stages with acute phase clinical samples. The DNA probe technology has already been used for detection and diagnosis of human and non human viral pathogens17. The present study describes the application of nucleic acid technology for the detection of chikungunya virus using a oligonucleotide probe labelled with biotin. The evaluation of this highly sensitive and specific DNA probe for clinical diagnosis of chikungunya virus is reported with a panel of 200 suspected acute phase human patient serum and CSF samples.
Material & Methods
Virus and cell lines: An Indian isolate of chikungunya virus CHIKV ISW HYD06 isolated from a febrile patient (Gen Bank accession number EF 210157) during 2006 epidemic in Hyderabad, Andhra Pradesh, India, was used in the present study. The virus was propagated in C6/36 cells (cloned cells derived from larvae of Stegomyia albopictus) and Vero (African green monkey, Cercopithicus aethiops kidney cells) in Eagle's minimum essential medium (EMEM) supplemented with 10 per cent FBS (foetal bovine serum) (Sigma, USA) obtained from National Center for Cell Science (NCCS), Pune, and was maintained in the Division of Virology, Defence Research and Development Establishment (DRDE), Gwalior, by regular sub-culturing at periodic intervals of 3-4 days. In addition, the four dengue virus serotypes (DEN-1, Hawaii; DEN-2, ThNH7/93; DEN-3, PhMH-J1-97; and DEN-4, SLMC 318), JE virus (P-20778), West Nile (WN) virus (Eg101) and Ross River (RR) virus were also used for checking the cross-reactivity. The synthetic gene constructs of three other related alphaviruses O’Nyong Nyong virus (ONN), Semlinki forest virus (SFV), and Sindbis virus (SINV) were also included in this study to check cross-reactivity due to non availability of respective alphaviruses.
Reverse transcription polymerase chain reaction (RT-PCR): The RNA was extracted from 140 μl of patients serum samples, CSF and infected culture supernatant using the QIAamp viral RNA mini kit (Qiagen, Germany) according to the manufacturers’ protocol. Finally, RNA was eluted in 50 μl of nuclease free water and used as template in RT-PCR. The following oligonucleotide primers were designed on the basis of available genome sequence of strain CHIKV ISW HYD06 (Gene Bank Accession No. 876190. To amplify the genomic region coding for nsP1, nsP3 and envelope (E1) gene, a set of primers were used (Table). The RT-PCR amplification of RNA was carried out using the Access quick one-step RT-PCR kit (Promega, Madison, WI, USA) in 50μl reaction volume containing 2x RT-PCR master mix, 2.5 U of AMV-RT and CHIKV specific primer pair sets.
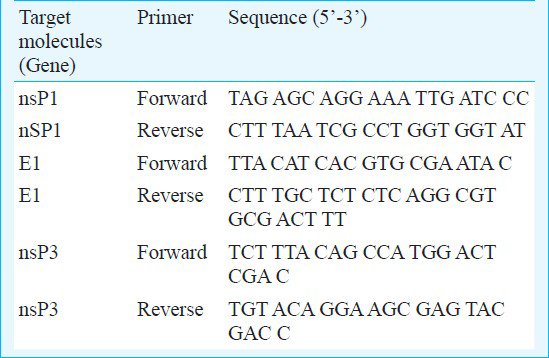
Preparation of oligonucleotide probe: Different genes (nsP1, nsP3 & E1) were targeted to generate the biotinylated probe. For that the purified fragment of different molecules followed by RT-PCR from standard strain of CHIKV was used for preparation of probe using Biotin DecaLabel DNA Labelling kit (MBI, Fermentas, USA). In brief, the method relies on priming of the polymerase reaction on the template DNA with random decanucleotide primers. The complementary strand is synthesized from the 3’ end of the primer using large fragment of DNA polymerase I, in the presence of nucleoside triphosphates, one of which is labelled with biotin. Biotin 11-dUTP is incorporated into the newly synthesized complementary DNA strand (cDNA).
Dot-blot hybridization: Complementary DNA (cDNA) made by reverse transcription of standard strain of CHIKV and clinical samples followed by reverse transcription method were spotted onto nylon membrane. Hybridization was perfomed as per the method of Bryan et al18 with slight modifications. Briefly, the spotted cDNA samples were air dried and cross-linked in UV cross linker for two min. The nylon membrane was subjected to prehybridization solution containing dextran sulphate (1 g), formamide (5 ml), SDS (sodium dodyl sulphate 100 mg) and NaCl sodium chloride (0.58 g) supplemented with denatured salmon sperm DNA, Sigma, USA (10 μg) for 2 h at 42°C. Hybridization was carried out in a solution containing 1mM EDTA, 5 per cent SDS and 100 ng/ml reaction mixture of labelled probe for 5 h at 42°C. After hybridization, the membrane was washed for 15 min with 1 × SSC containing 10 per cent SDS at room temperature (RT), and then for 15 min at 50 °C with 20X SSC solution containing 10 per cent SDS.
Colour development of biotinylated probe: After hybridization, the nylon membrane was blocked with 3 per cent BSA and incubated with streptavidin HRP conjugate (1: 3000 dilution in 3 % BSA) in buffer A (0.1 M Tris/HCl pH 7.5, 0.1 M NaCl, 2 mM MgCl2, 0.05% Triton X-100) for 30 min at room temperature with end to end shaking19. The membrane was washed twice for 7 min at RT in buffer A, then twice for 7 min at RT in buffer B (0.1 M Tris/HCl pH 9.5, 0.1 M NaCl, 50 mM MgCl2,). Following washing, the membrane was developed using a colour development solution containing DAB chromogen. The reaction was stopped in deionized water and air-dried for storage. Non specific background was eliminated by optimizing the concentration of the streptavidin alkaline phosphates conjugate, including Denhardt's solution in the pre-hybridization and hybridization buffers, incubation the colour development step in the dark, and limiting the incubation time to under 1 h20.
Sensitivity and specificity of the probe: The sensitivity of the biotinylated probe under optimal conditions was determined by hybridizing the probe to dots containing dilutions of cDNA ranging from 107 to 0.1 copies of standard strain of chikungunya virus. Ross River virus (RRV) and SLE were used as negative controls. In addition, the specificity of the CHIKV E1 gene specific DNA probe was established with a panel of 20 healthy human serum samples as well as RRV and SLE virus culture supernatant, 30 confirmed dengue cases from an outbreak in 2004 in India and 25 serological positive samples of JE patients were also included in this study. In absence of the virus, the synthetic gene constructs of O’Nyong Nyong virus (ONN), Semliki forest virus (SFV), and Sindbis virus (SINV) were also used to check cross-reactivity.
Evaluation of probe: A total of 200 clinical samples (comprising 160 serum and 40 CSF samples) collected from patients suspected to have chikungunya infection admitted with clinical diagnosis of polyarthritis in medicine wards at Central India Institute of Medical Sciences (CIIMS), Nagpur and District surveillance unit, Kasturba Medical College (KMC), Dakshin Karnataka, Mangalore, in 2005 and 2007 were used in this study. Patients’ consent and approval were obtained from the Institutional Ethics Committees of CIIMS, Nagpur and KMC, Mangalore. The acute phase samples were collected during the period between days 1 to 7 after the onset of symptoms. All the samples were transported to the laboratory under strict cold chain and stored at -80°C till further investigation. In order to check the cross-reactivity, serum samples from 30 confirmed dengue patients were also included due to similar clinical signs and symptoms. In addition, a panel of 20 serum samples collected from healthy individuals from Gwalior was also included as negative controls. The comparative evaluation of the CHIKV E1 specific DNA probe was carried out in parallel to CHIKV E1 gene specific SYBR Green I based real-time RT-PCR21.
SYBR green-I based real time RT-PCR: DNA probe positive serum and CSF samples were further analyzed by SYBR Green-I based one-step real time quantitative RT-PCR by targeting the E1 gene in MX 3000P quantitative PCR system according to the protocol described by Santhosh et al21. Briefly, RNA was extracted by QiaAmp viral mini kit and was subjected to real time RT-PCR by using ‘Brilliant SYBR Green Single-Step QRT-PCR Master Mix’ (Stratagene, Cedar Creek, Texas, USA), in a 25 μl reaction mixtures containing 12.5 μl of 2x reaction mix, 0.4 μl of reference dye (ROX), 1 μl (10 pmol) of each forward and reverse primers, 1 μl of RNA, 0.1 μl of reverse transcriptase, 9.0 μl of nuclease free water. Suitable controls without template and primer kept alongside and buffer controls were also included in the tests. Following amplification, a melting curve analysis was performed to verify the authenticity of the amplified product by its specific melting temperature (Tm).
Indirect ELISA: An in-house developed indirect microplate IgM ELISA was performed using the purified recombinant protein of chikungunya virus. The purified envelope (E2) protein was diluted to 300 ng/μl in 0.1 M carbonate buffer, pH 9.6, and used for coating in 96-well microtiter plate (100 μl/well) and incubated at 37°C for 2 h. The coated wells were washed once with 1 x phosphate buffer saline (PBS) and then blocked with 3 per cent BSA in 1x PBS overnight at 4°C. Following washing as above 100 μl of CHIK specific serum samples at a dilution of 1:100 made in 1x PBS were added in the wells and incubated for 1 h at 37°C. The plate was incubated with anti-human IgM-HRP conjugate (Sigma, USA) (1:3000 dilutions in 3% BSA) followed by washing steps. Reaction was developed with 100 μl of tetra methyl benzidine (TMB, Sigma, USA) substrate and kept at RT for 5 min. The peroxidase reaction was terminated with 100 μl of 1 N H2SO4 and the absorbance was read at 490 nm in a ELISA reader (Biotep, USA)23.
Sandwich ELISA: Double antibody sandwich ELISA test system was optimised employing the rabbit hyper immune sera (HIS) and mouse Mab against CHIK E2 specific as capture and detector antibodies, respectively. The wells of ELISA plate were coated with 100 μl of 1:1000 dilution of rabbit HIS and incubated at 4°C overnight. Following washing with 1x PBS, the wells were blocked with 3 per cent BSA followed by incubation at 37 °C for 2 h. After washing with 1x PBS, 100 μl of 1:100/ 1: 10 dilution of patient serum/CSF samples were added in the wells and incubated at 37°C for 1 h. The plate was incubated with 100 μl of 1:500 dilution of high affinity mouse Mab followed by washing steps. After washing, the wells were incubated with anti-mouse HRP conjugate at a dilution of 1:2000 made in 3 per cent BSA. Reaction was developed with 100 μl of TMB substrate and kept at room temperature for 5 min. The peroxidase reaction was stopped with 100 μl of 1 N H2SO4 and the absorbance was read at 490 nm in a ELISA reader23.
Results
Dot-blot hybridization: Out of three different targeted molecules, E1 labelled product was found to be highly specific. A 500 bp fragment encoding E1 gene was amplified by RT-PCR using employing CHIKV E1 gene specific primers. This amplified product was purified and labelled with biotin and utilized for hybridization (Fig. 1). The E1 targeted biotinylated probe was specifically hybridized with complementary sequence of the cDNA followed by reverse transcription of standard strain of chikungunya virus. An optimal assay condition was established wherein no signal was observed with other closely related members of alpha and flaviviruses family viz. RRV, SLE, dengue, and JE. Other targeted molecules nsP1 and nsP3 showed cross-reactivity with other alphavirus (RRV).
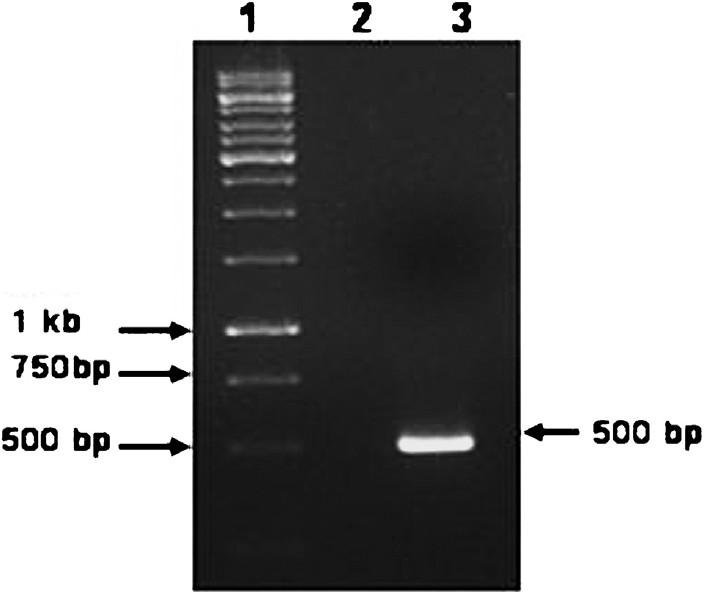
- Agarose gel electrophoresis showing the amplification of the expected 500 bp PCR product. Lane 1 - Molecular weight marker (1 Kbp DNA ladder); Lane 2- Negative control (No template); Lane 3 - Positive control (Novel ECSA genotype).
Sensitivity and specificity of DNA probe: The sensitivity of the CHIKV E1 specific DNA probe was determined by testing serial 10-fold dilutions of different concentration ranging from 107 to 0.1 copies of standard strain of chikungunya virus. The detection limit of the CHIK E specific probe was found to be 1 copy number/ml (Fig. 2). The specificity of the CHIKV E1 specific DNA probe was established by checking the cross-reactivity with other closely related members of alphavirus group such RRV and SLE virus culture supernatant as well as plamid DNA of synthetic gene constructs of O’Nyong Nyong virus (ONN), Semlinki forest virus (SFV), and Sindbis virus (SINV). In addition, the CHIKV E1 gene specific DNA probe did not show any positivity with 30 dengue and 25 JE confirmed human patients acute phase serum samples having similar clinical picture. Further specificity of the chikungunya specific DNA probe was also established by screening 20 serum samples from apparently healthy individuals where in no false positive reaction was observed.

- The sensitivity of the CHIKV E1 gene specific DNA probe as determined with serial 10-fold dilutions ranging from 107 to 0.1 copy number of standard strain of chikungunya virus. The detection limit of the DNA probe was found to be 10 copy number indicated by the appearance of faint dot. No dot was appeared in RRV and SLE viruses.
The efficacy of CHIKV E1 gene specific DNA probe in picking up chikungunya positive samples was compared with in-house developed sandwich ELISA for antigen detection and real time RT-PCR employing CHIKV specific E1 gene primers. The amplification and dissociation plots showed the CHIKV specific melting temperature (Tm = 81.6 °C) as revealed by SYBR Green I- based one-step quantitative real-time PCR using E1 gene specific primer sets (Fig. 3). The comparative evaluation with SYBR Green I based real-time RT-PCR revealed 96 per cent concordance with a sensitivity and specificity of 96 and 95 per cent, respectively. The positive and the negative predictive values of the E1 gene specific DNA probe for picking up Chikungunya virus genomic RNA were found to be 94 and 97 per cent. All the DNA probe positive samples were also confirmed by cycle sequencing (data not shown).
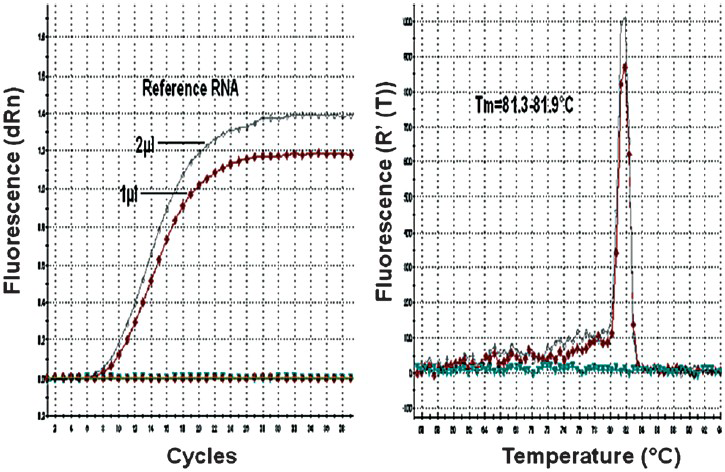
- Real-time kinetics of CHIKV E1 gene specific SYBR Green I based real-time RT-PCR showing the amplification and dissociation curve. A. Amplification plot. B. Melting curve analysis depicting dissociation plot.
Of the 200 clinical samples (160 serum and 40 CSF), 71 serum and nine CSF samples were showed positive signal in the form of brown colour dot with chikungunya E1 gene specific DNA probe (Fig. 4). The positivity for chikungunya virus among the serum and CSF samples was found to be 45 and 22 per cent, respectively. No cross-reactivity was observed with infected culture fluid of RRV and SLE virus as well as healthy human serum samples. In addition, patient serum with confirmed dengue and JE also did not reveal any hybridization.
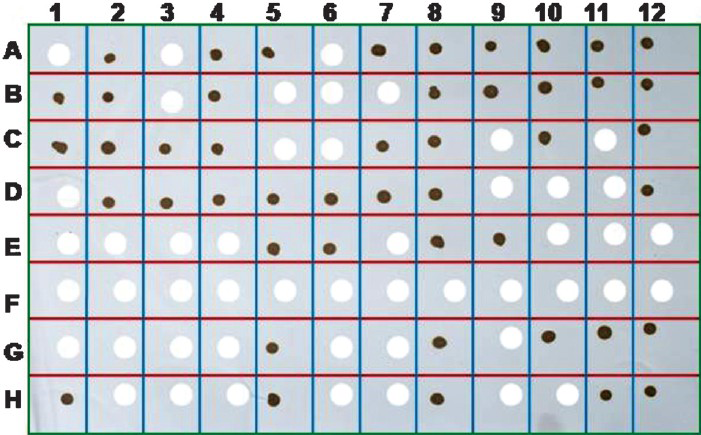
- The dot blot analysis of the human patient acute phase serum and CSF samples for the presence of genomic RNA of chikungunya virus employing the CHIK E 1 specific biotinylated DNA probe. The appearance of brown colour dot following colour development with DAB chromogen confirmed the presence of chikungunya virus. The blank space shows negative result indicating the absence of chikungunya virus.
The DNA probe system reported here was found to be highly sensitive and specific as it did not miss any of the real time RT-PCR positive cases and also did not reveal any false positivity among the negatives.
Sandwich ELISA: Of the 200 clinical samples (160 serum and 40 CSF), 65 serum and seven CSF samples showed positivity for sandwich ELISA. The positivity for chikungunya virus among the serum and CSF samples was found to be 40 and 18 per cent, respectively. However, some of the sandwich ELISA negative cases were found to be positive by DNA probes method (data not shown). This is because the higher sensitivity of DNA probe assay. A sample was considered positive if the optical density (OD) was greater than twice the mean value of the negative controls.
Indirect IgM ELISA: Of the 200 clinical samples, 45 (22 %) were positive for IgM antibodies by indirect ELISA employing E2 recombinant antigen. The cut-off value was calculated based on twice the average OD value of confirmed negative samples. Commercial mu Capture ELISA involves 3 steps and takes 4 h to complete the assay. Whereas our in-house developed recombinant based indirect IgM ELISA takes 1 h 30 min to complete the test. In-house IgM ELISA was validated on a large number of samples and it revealed 95 per cent correlation with commercially available mu capture ELISA.
Discussion
Chikungunya virus infections are rarely fatal and generally do not require admission to a hospital. Despite the fact that CHIKV resurgence is associated with an epidemic of unprecedented magnitude, only a few serological and molecular diagnosis tools are available. However, it is important to identify and quantify this infection for clinical diagnosis as well as epidemiological studies.
Currently, the diagnosis of CHIKV infection is accomplished through either virus isolation or genomic detection by RT-PCR13. Serum IgM antibodies against CHIK V can be detected from patients as early as 3 to 5 days after the onset of fever and generally persist for 30-90 days, although detectable levels may be present up to eight months post-infection14.
The DNA probe assay reported here also used amplification of chikungunya virus nucleic acids in cell culture, analogous to amplification of antigens for serological tests. However, viral mRNA appears earlier in infected cells than its corresponding protein and can be rapidly extracted for analysis. In addition, antibodies, particularly monoclonal antibodies, are expensive and time consuming to develop, while DNA oligonucleotide probes can be inexpensively synthesized.
The currently available commercial kits are based on IgM antibody detection and thus have less significance for early diagnosis of the patients. However, all nucleic acid amplification methods have several disadvantages, including requiring either a high-precision instrument for amplification or an elaborate, complicated method for the detection of amplified products.
The present study describes the standardization and evaluation of DNA probe for detection of chikungunya virus. In addition, the test system can also be useful for detection of viral genome in the infected culture supernatant and thus can be an integral part of quality control of chikungunya virus production in the laboratory. The present results were compared with those of SYBR Green I based quantitative RT-PCR assay which has several advantages over the previously reported TaqMan assay such as, less expensive, simple in its manipulation than TaqMan assay. The validation of this assay with clinical samples from epidemic further confirmed its applicability in clinical setting. Comparative evaluation of DNA probe with SYBR Green real-time RT-PCR revealed more than 95 per cent concordance. The applicability of this DNA probe was also hecked for other genotypes including the S-27 (prototype strain of African genotype), and Asian genotype as well as the novel ECSA genotype with and without A226V mutation (data not shown). The higher sensitivity and specificity of the DNA probe suggest that this probe assay is potentially useful as a diagnostic intermediate in the screening of CHIKV suspected samples. Cross-reactivity with other members of alphaviruses and flaviviruses was significantly reduced while identifying CHIKV infection by DNA probe and was found to be non reactive with all these serum samples thus establishing its high specificity.
Biotinylated oligonucleotide probes are well studied for in situ hybridization using tissue sections from infected animals192024. This technique, in combination with PCR has significant application for research into the carrier state of chikungunya virus, as it has the potential to detect nucleic acids of viruses in a latent state.
The potential advantages of these DNA probe assays in the diagnosis of infectious diseases include rapid detection and identification of infectious agents, the ability to screen selected specimens using batteries of probes, and the detection of nonviable or difficult-to-culture organisms. The DNA probe assay reported in the present study has application in the molecular diagnosis of chikungunya infection. The overall data obtained from the extensive evaluation with reasonably good number of acute phase serum and CSF samples indicated that the DNA probe reported in this study is highly specific and sensitive for the detection of chikungunya virus. The assay is also useful in laboratory for detection, quantification and quality control of viral antigen in cell culture supernatant. This may be attributed to the higher sensitivity of the biotinylated assay that can pick up only a few copies of the viral genome whereas antigen detection requires a high level of viraemia which may not be the case always19.
In conclusion, the overall higher sensitivity and specificity of the reported biotinylated DNA probe offers a promising economical assay format for molecular diagnosis and surveillance of chikungunya virus.
Acknowledgment
The authors thank Dr R. Vijayaraghavan, Director, Defence Research and Development Establishment (DRDE), Ministry of Defence, Government of India, for his support, constant inspiration and providing the necessary facilities for this study. The authors also thank Dr G.M. Taori, Director, CIIMS, Nagpur, and District surveillance unit, Dakshin Karnataka, Mangalore, for providing the clinical samples.
References
- Re-emergence of Chikungunya and O’nyong-nyong viruses: evidence for distinct geographical lineages and distant evolutionary relationships. J Gen Virol. 2000;81:471-9.
- [Google Scholar]
- Alphaviruses. In: Fields BN, Knipe DM, Howley PM, eds. Fields virology (3rd ed). Philadelphia: Lippincott-Raven Publishers; 1996. p. :843-98.
- [Google Scholar]
- An epidemic of virus disease in Southern Province, Tanganyika Territory, in 1952-53. I. Clinical features. Trans R Soc Trop Med Hyg. 1955;49:28-32.
- [Google Scholar]
- Epidemic resurgence of chikungunya virus in democratic Republic of the Congo: identification of a new central African strain. J Med Virol. 2004;74:277-82.
- [Google Scholar]
- The alphaviruses: gene expression, replication and evolution. Microbiol Rev. 1994;58:491-562.
- [Google Scholar]
- Genome microevolution of chikungunya viruses causing the Indian Ocean outbreak. PLoS Med. 2006;3:e263.
- [Google Scholar]
- Changing patterns of chikungunya virus: re-emergence of a zoonotic arbovirus. J Gen Virol. 2007;88:2363-77.
- [Google Scholar]
- Chikungunya outbreaks caused by African genotype, India. Emerg Infect Dis. 2006;12:1580-3.
- [Google Scholar]
- East Central South African genotype as the causative agent in reemergence of chikungunya outbreak in India. Vector Borne Zoonotic Dis. 2007;7:519-27.
- [Google Scholar]
- Comparative full genome analysis revealed E1: A226V shift in 2007 Indian chikungunya virus isolates. Virus Res. 2008;135:36-41.
- [Google Scholar]
- Specific detection of chikungunya virus using a RT-PCR /nested PCR combination. J Vet Med B Infect Dis Vet Public Health. 2002;49:49-54.
- [Google Scholar]
- Rapid detection of Chikungunya virus in laboratory infected Aedes aegypti by reverse-transcriptase polymerase chain reaction (RT-PCR) Trop Biomed. 2005;22:149-54.
- [Google Scholar]
- Evaluation of a rapid assay for detection of IgM antibodies to Chikungunya. Southeast Asian J Trop Med Public Health. 2010;41:92-6.
- [Google Scholar]
- Detection of alphaviruses in a genus-specific antigen capture enzyme immunoassay using monoclonal antibodies. J Clin Microbiol. 1991;29:131-7.
- [Google Scholar]
- Molecular and serological diagnosis of Chikungunya virus infection. Pathol Biol (Paris). 2007;55:490-4.
- [Google Scholar]
- Development of a TaqMan RT-PCR assay without RNA extraction step for the detection and quantification of African Chikungunya viruses. J Virol Methods. 2005;124:65-71.
- [Google Scholar]
- Rapid and real-time detection of Chikungunya virus by reverse transcription loop-mediated isothermal amplification assay. J Clin Microbiol. 2007;45:351-7.
- [Google Scholar]
- Diagnosis of clinical samples with synthetic oligonucleotide hybridization probes. In: Leive L, ed. Microbiology. Washington, D.C: American Society of Microbiology; 1986. p. :113-6.
- [Google Scholar]
- Development of biotinylated DNA probe for detection and identification of infectious hematopoietic necrosis virus. Dis Aquat Organ. 1991;11:57-65.
- [Google Scholar]
- A method for biotinylating oligonucleotide probes for use in molecular hybridizations. DNA. 1986;5:333-7.
- [Google Scholar]
- Development and evaluation of SYBR Green I- based one-step real-time RT-PCR assay for detection and quantification of chikungunya virus. J Clin Virol. 2007;39:188-93.
- [Google Scholar]
- Enzyme linked immunosorbent assay (ELISA). Quantitative assay of immunoglobulin G. Immunochemistry. 1971;8:871-4.
- [Google Scholar]
- Detection of chikungunya virus antigen in Aedes albopictus mosquitoes by enzyme-linked immunosorbent assay. J Virol Methods. 1985;12:279-85.
- [Google Scholar]
- Synthetic oligonucleotide probes for the detectión of human papilloma viruses by in situ hybridisation. J Virol Methods. 1988;20:239-49.
- [Google Scholar]






