Translate this page into:
Consistency of standard laboratory strain Mycobacterium tuberculosis H37Rv with ethionamide susceptibility testing
Reprint requests: Dr. Ranjani Ramachandran, Scientist “E”, Department of Bacteriology, National Institute for Research in Tuberculosis (ICMR), Chetput, Chennai 600 031, India e-mail: ramachandranr@searo.who.int
-
Accepted: ,
This is an open-access article distributed under the terms of the Creative Commons Attribution-Noncommercial-Share Alike 3.0 Unported, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
This article was originally published by Medknow Publications & Media Pvt Ltd and was migrated to Scientific Scholar after the change of Publisher.
Abstract
Drug susceptibility pattern of standard Mycobacterium tuberculosis strain H37Rv showed discrepancy in minimum inhibitory concentration method for ethionamide and consistent results were obtained for the other second line drugs namely, kanamycin and ofloxacin. It is, therefore, necessary to revisit the susceptibility testing method for ethionamide for effective clinical management of patients with drug resistant tuberculosis.
Keywords
Drug susceptibility testing
ethionamide
Mycobacterium tuberculosis H37Rv
The susceptibility pattern for the standard laboratory strain of Mycobacterium tuberculosis H37Rv was found to vary with respect to second line drugs, kanamycin (KAN), ethionamide (ETO) and ofloxacin (OF) currently being used under the Revised National Tuberculosis Control Programme1. As second line treatment was initiated in India during 2009, it becomes necessary to check the efficiency of the standard strain from time to time. Conventional minimum inhibitory concentration (MIC) and proportion sensitivity test (PST) methods were employed for drug susceptibility testing (DST) following standard procedures2. The aim of this study was to check the consistency of the standard strain used as laboratory control in conventional DST methodology. At 35 different time points spanning one year time period, the strain was used as control during DST procedures.
Susceptibility testing for ETO by MIC method indicated discordant value as high as 28.6 per cent (10/35) and 5.7 per cent (2/35) by PST method (Table). DST of ETO is known to be dynamic and it is tedious to deduce an accurate method for detecting resistant and sensitive strains3. One probable reason is that, MIC method uses a high concentration of inoculum (4 mg/ml) than the PST method (1 mg/ml). Various other reasons for the development of inconsistency by MIC method may be primarily technical such as selection of representative clonal population, proper and appropriate preparation of suspension without any clumping, accurate inoculation (10 μl/slope) and preparation of media with the correct drug concentration. To a limited extent, presence of borderline/intermediate population may induce some discrepancy in DST procedures. This phenomenon is inevitable in bacteriostatic drugs such as ETO4. The MIC level is very near to its absorption maxima; hence, inconsistent DST with respect to ETO especially by MIC method is well expected. Using PST method for DST for ETO, the consistency level was high compared to MIC method. Only on two occasions H37Rv was found to be resistant. In PST method, inoculum is diluted and then mixed thoroughly to get a suspension making errors related to technique minimal.
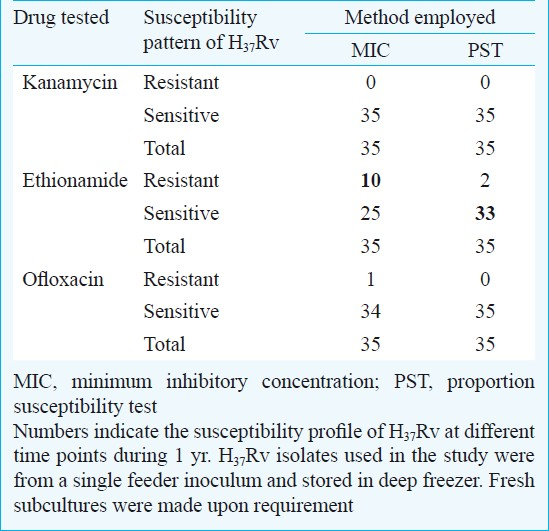
ETO is a thermolabile drug and there is a speculation that the dynamic susceptibility pattern observed in solid culture DST may be the result of drug deterioration either during media preparation or during incubation in DST procedures3. Problems associated with drug deterioration are expected in PST method because of the longer incubation period (6 wk) than MIC (4 wk). In contrast, the results showed a consistent susceptibility profile using PST method than MIC method and are in line with the earlier report3. This can be attributed to the inoculum size which is four times more in MIC method compared to PST. High inoculum may introduce more clumps (if not prepared stringently) and as a result an increased inoculum is being delivered onto the drug containing slope. This may not be the cause at all instances as the technical personnel had been trained in DST methodology and monitored periodically.
The advantage in this study was that H37Rv isolates used were primary cultures from a single feeder inoculum and stored in deep freezer. Fresh subcultures were made upon requirement. Hence, it eliminates the effect of repeated sub-culturing that could be a cause for discrepant results. This is an indication for urgent requirement of an effective method to define DST for ETO. If the existing methods for susceptibility testing are unable to provide perfect result, then modifying or optimizing the existing techniques either in the methodology or interpretation can be undertaken. When the standard strain was used as control in liquid culture systems (BACTEC460TB and MGIT960), the results were consistent and designated as susceptible at their critical concentration defined by World Health Organization (unpublished data). This indicates that there is a defect in the existing DST methodology using solid media.
One more reason for the discrepant results for ETO may be due to variation in standard strain used in the laboratory. It is believed that H37Rv maintained in different laboratories across the world has a minimum level of genetic variation (<0.01%)5. Hence, H37Rv strains maintained in laboratory under controlled environment might not induce any spontaneous mutations that are usually expected among clinical strains in due course of evolution. But this theory was proved wrong by a recent study6 where distinct polymorphisms were observed in standard strains maintained across six laboratories. The authors have concluded that even under controlled environment, H37Rv is evolving evidenced by in vitro accumulation of genetic differences during serial passaging of cultures6. Therefore, it becomes necessary to perform genetic screening of the standard strain at regular time intervals to assess the nature of H37Rv maintained to exclude genomic variation as a cause for inconsistent DST results.
The results for the most important second line drugs, KAN and OF were in agreement by both DST methods. The drug KAN showed susceptible phenotype at all time points by both methods. Ofloxacin, at a single time point showed resistant phenotype by MIC method and showed concordant results by PST method (Table). The reason can be attributed to ability of the drug to provide a clear demarcation between resistant and susceptible strains. Such a validation proves to be important when detection of extensively drug resistant TB (XDR-TB) is required. Since, the laboratory strain does not show much of variation for these drugs by both methods, either MIC or PST method can be used for determining susceptibility profile to KAN and OF. Similar concordant results by both MIC and PST methods were observed with clinical isolates in the laboratory (data not shown).
It is, therefore, necessary for all laboratories involved in mycobacteriology procedures to have such check points on their standard strain H37Rv from time to time as one of the quality assurance measures. If facilities are available, sequencing of randomly selected cultures and discrepant strains to identify the polymorphism can indicate the exact reason for such discrepancy.
Acknowledgment
The financial assistance provided by World Health Organization (WHO) through NIH/USAID and Indian Council of Medical Research (ICMR), New Delhi, for infrastructure facilities is acknowledged. The first author (LR) thanks ICMR for financial support as SRF.
References
- RNTCP DOTS-Plus Guidelines. 2010. Central TB Division. Available from: http://www.tbcindia.org
- [Google Scholar]
- Advances in techniques of testing mycobacterial drug sensitivity, and the use of sensitivity tests in tuberculosis control programmes. Bull World Health Organ. 1969;41:21-43.
- [Google Scholar]
- Comparison of methods for testing the sensitivity of Mycobacterium tuberculosis to ethionamide. Tubercle. 1966;47:250-61.
- [Google Scholar]
- Variation among genome sequences of H37Rv strains of Mycobacterium tuberculosis from multiple laboratories. J Bacteriol. 2010;192:3645-53.
- [Google Scholar]






