Translate this page into:
Complete genome sequence of two genotype III Japanese encephalitis virus isolates from West Bengal, India
Reprint requests: Dr Shyamalendu Chatterjee, GB4, 1st floor, ID & BG Hospital, 57, Dr. S.C. Banerjee Road, Beliaghata, Kolkata 700 010, West Bengal, India e-mail: shyamalendu.chatterjee1@gmail.com, taraphdar.debjani@gmail.com
-
Received: ,
This is an open-access article distributed under the terms of the Creative Commons Attribution-Noncommercial-Share Alike 3.0 Unported, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
This article was originally published by Medknow Publications & Media Pvt Ltd and was migrated to Scientific Scholar after the change of Publisher.
Abstract
Background & objectives:
Japanese encephalitis (JE), caused by a mosquito-borne virus JE virus (JEV), is a serious health problem in West Bengal, India. In this study, we report the complete genome sequence of two JEV isolates from West Bengal. The amino acid and nucleotide sequence homology was compared with other Indian strains.
Methods:
Two JEV isolates (IND-WB-JE1 and IND-WB-JE2) obtained in 2008 and 2010, respectively, from two districts of the State of West Bengal, respectively were analyzed for genetic variations by sequencing the 10934 bp whole genome of the virus. Of these two districts, one was covered under JE vaccination programme in 2007.
Results:
Phylogenetic analysis showed that both the isolates belonged to the genotype III. A total of 16 mutations were identified in the two isolates studied with respect to Vellore P20778 strain. One unique mutation A3215S was only found in IND-WB-JE2 isolate, but not in the isolate IND-WB-JE1. These two isolates showed maximum homology with P20778 strain of India.
Interpretation & conclusions:
This study reports on complete gene based phylogenetic analysis of JEV isolates from the State of West Bengal. It was evident from the results that JEV was still under circulation in both vaccine covered and not covered districts of West Bengal.
Keywords
Genetic variations
Japanese encephalitis virus
molecular characterization
mutations
West Bengal
Japanese encephalitis (JE), a mosquito-borne life threatening viral disease caused by Japanese encephalitis virus (JEV), is endemic in a large portion of Asia and South East Asia12. Every year, 68,000 clinical cases and 20,400 deaths occur due to the disease3.
JEV is the member of the genus Flavivirus of the Flaviviridae family4. It has a single stranded, positive sense RNA genome, which is capped at the 5’ end. The genome is approximately 11 kb in length and contains one open reading frame (ORF). The 5’one-third of the ORF encodes three viral structural proteins, namely, capsid (C), membrane (M) and envelope (E), while the 3’ two-third region encodes non-structural proteins, namely, NS1, NS2A, NS2B, NS3, NS4A, NS4B and NS55.
JEV was first isolated from Japan in 19356. In India, the first case of JE was recorded from Vellore, Tamil Nadu in 19557. In West Bengal, the first major outbreak of JE was reported from the districts of Bankura and Burdwan in 19738; about 700 cases and 300 deaths occurred in that outbreak. From 1978 to August 2013, 107,552 cases and 34,461 deaths due to JEV infection were recorded from various parts of India69. According to the data of the National Vector Borne Disease Control Programme (NVBDCP), till August 2013, a total of 209 cases and 18 deaths occurred due to JEV infection in the State of West Bengal after vaccination9.
In India, during 2006-2009, children in the age group of 1-15 yr were vaccinated with live attenuated JE vaccine of SA-14-14-2 strain, manufactured by Chengdu institute of Biological Products, Chengdu, China10. Most affected areas of some JE endemic States were selected. It included seven districts of Uttar Pradesh, two districts of Assam and one district each from West Bengal and Karnataka11. Between 2007 and 2009, vaccination programme was initiated in Andhra Pradesh, Bihar, Haryana, Maharashtra, Tamil Nadu, Kerala and Goa11.
In West Bengal, JE is still a public health problem, especially among children12. The present study was carried out to detect the mutations in the genome of the JEV that are circulating in West Bengal, India. Complete genome sequences (verified) of only five isolates from India are available in GenBank, of which one is from equine13. We determined the full-genome sequences of two JEV isolates from two districts of West Bengal, one from a district covered under JE vaccination in 2007 and another not covered. The homology of the nucleotide and amino acid sequences of the whole genome was precisely compared with other Indian JEV strains.
Material & Methods
This study was conducted in the laboratory of ICMR Virus Unit, Kolkata, West Bengal, India, during 2008-2010. JEV P20778 strain (GenBank Accession No. AF080251) was obtained from the National Institute of Virology (NIV), Pune, India, and was used as a positive control throughout the study.
Preparation of virus stock: The lyophilized virus was reconstituted with miliQ water and was seeded at multiplication of infection (MOI) equal to 1 on the 90 per cent monolayer of Vero cell lines. The culture was observed regularly for the appearance of cytopathic effect (CPE) up to 7-8 days. On appearance of the CPE, virus was harvested by repeated freezing and thawing of the cultures followed by centrifugation at 3000 g for 15 min at 4°C to remove the cell debris and the supernatant was used as positive control for reverse transcriptase polymerase chain reaction (RT-PCR) or stored at -80°C for future use.
Isolation of virus: For the purpose of sequencing of the circulating strains, attempts were made to isolate the virus. Two JEV isolates, IND-WB-JE1 obtained in 2008 from Malda district of West Bengal (no vaccinated done) and IND-WB-JE2 obtained in 2010 from Birbhum district (vaccine covered under JE vaccination programme in 2007) were used in this study.
These samples were adsorbed in the 80-85 per cent confluent monolayers of Vero cell lines for two hours in a 6-well tissue culture plates (Nunc, Denmark) at 37°C as per the standard protocols14. Infected vero cells were incubated at 37°C till the appearance of CPE or up to five days of post-inoculation. After the appearance of CPE, the infected tissue culture fluid was centrifuged at 3000 g for 15 min at 4°C. The supernatant of the tissue culture fluid (STF) was used for further studies. Non-infected Vero cell culture was used as negative control.
RNA was isolated from the STF by using QIAamp® RNA viral kit (Qiagen, GmbH, Hilden, Germany). The purified RNA was used as template for cDNA synthesis using SuperScript™ III first-strand synthesis system (Invitrogen, USA) with reverse JWR1 5’-AGATCCTGTGTTCTTCCTCACCACCA-3’ primer according to the manufacturer's instructions. Subsequent PCR amplification was carried out to amplify the overlapping viral genomic cDNA fragments with different oligonucleotide primers (self designed, ICMR, Virus Unit, Kolkata, India) by using Platinum® Taq DNA Polymerase High Fidelity (Invitrogen, USA) (Table I). The PCR products were analyzed in 0.8 per cent agarose gel electrophoresis.
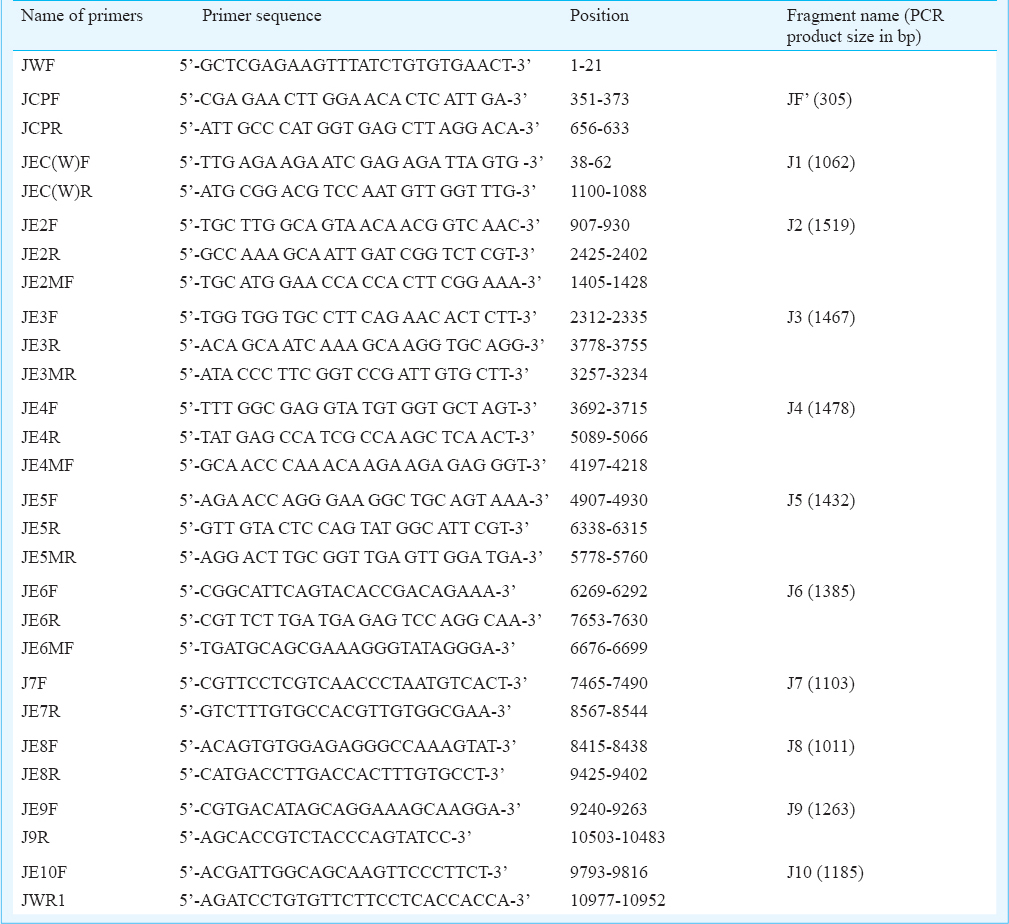
Sequencing and phylogenetic analysis: The amplified products were purified by Qiagen Gel Extraction Kit and were sequenced by BigDye Terminator Cycle Sequencing Ready Reaction Kit (Applied Biosystems, USA) as per the manufacturer's instructions. The products were analyzed using an automated DNA sequencer, 3130XL Genetic Analyzer (Applied Biosystems).
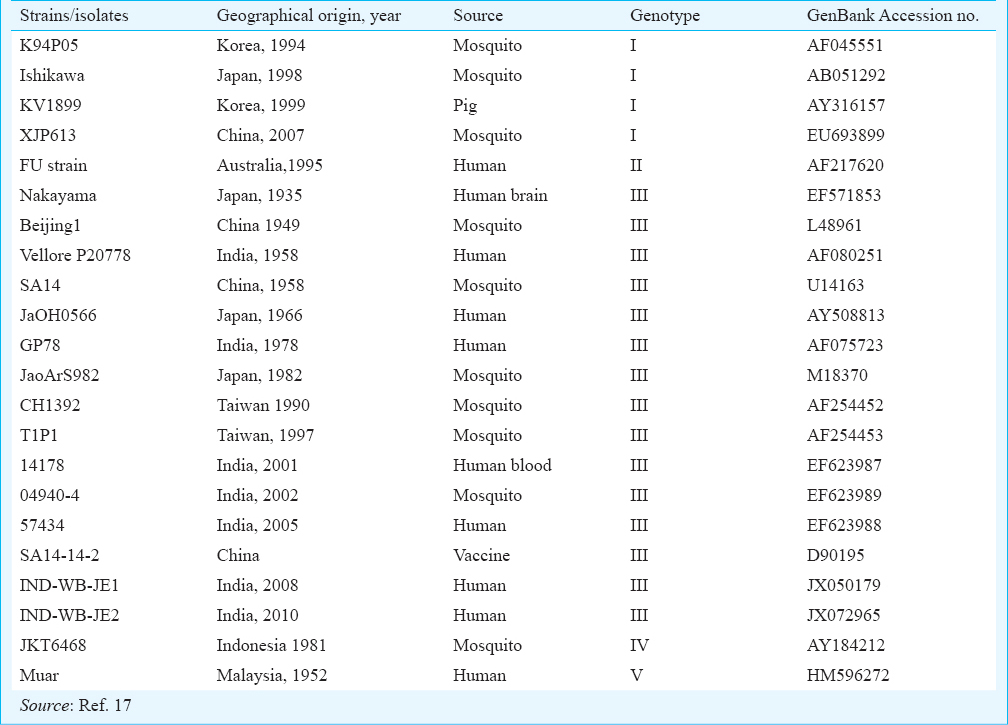
The data were submitted to the GenBank for the accession numbers. The raw data (ABI file) were converted in to the ‘txt’ format by the software ‘Bioedit’15 for further analysis. The Neighbour Joining (NJ) phylogenetic tree was constructed based on the 20 JEV reference sequences available in GenBank using the software MEGA 4.116. The source, year of collection and GenBank accession numbers of the studied isolates are given in Table II. The reliability of different phylogenic groupings was evaluated with the bootstrap test (500 bootstrap replications) available in MEGA. This was followed by the calculation of genetic distances using Kimura two-parameter (K2P) model16.
Results
RT-PCR result showed specific bands at every required position in the two isolates on 0.8 per cent agarose gel. These samples also produced prominent CPE in tissue culture system. The IND-WB-JE1 was isolated in 2008 from the district of Malda, where no vaccination has yet been done, and the other one, IND-WB-JE2 was isolated from the district of Birbhum (vaccine covered district) in 2010. The full-length assembled JEV genome consisted of 10,934 base pairs which corresponded to the positions 1-10,934 of the reference sequence (Vellore P20778).
The two isolates from West Bengal i.e. IND-WB-JE1 (GenBank: JX050179) and IND-WB-JE2 (GenBank: JX072965) showed maximum similarity (99%) with the Vellore P20778 strain (GenBank: AF080251), followed by 97 per cent nucleotide similarity with Nakayama (GenBank: EF571853), Beijing-1 (GenBank Acc. no L48961), SA-14 (GenBank Acc. no U14163), JaGAr01(GenBank Acc. no AF069076), CH1392 (GenBank Acc. no AF254452) and SA-14-14-2 (GenBank Acc. No D90195) and 96 per cent nucleotide similarity with GP78 (GenBank Acc. no AF075723), 014178 strain (GenBank Acc. no EF623987), 04940-4 strain (GenBank Acc. no EF623989), 057434 strain (GenBank Acc. no EF623988), JaoArS982 (GenBank Acc. no M18370) and JaOH0566 (GenBank Acc. no AY508813). These two West Bengal JEV isolates also showed 99 per cent nucleotide similarity with each other.
Phylogenetic analysis based on complete genome of the two West Bengal isolates (IND-WB-JE1 and IND-WB-JE2) and 20 other isolates including five Indian isolates available in GenBank, indicated almost close clustering of the West Bengal isolates in genotype III (Figure). However, the West Bengal isolates formed separate cluster with 100 per cent bootstrap support and showed very close clustering with Vellore-P20778 strain. The findings of this study corroborated with that of a previously reported study, where the Vellore strain and GP-78 strain of India formed separate clusters in the phylogenetic tree18.
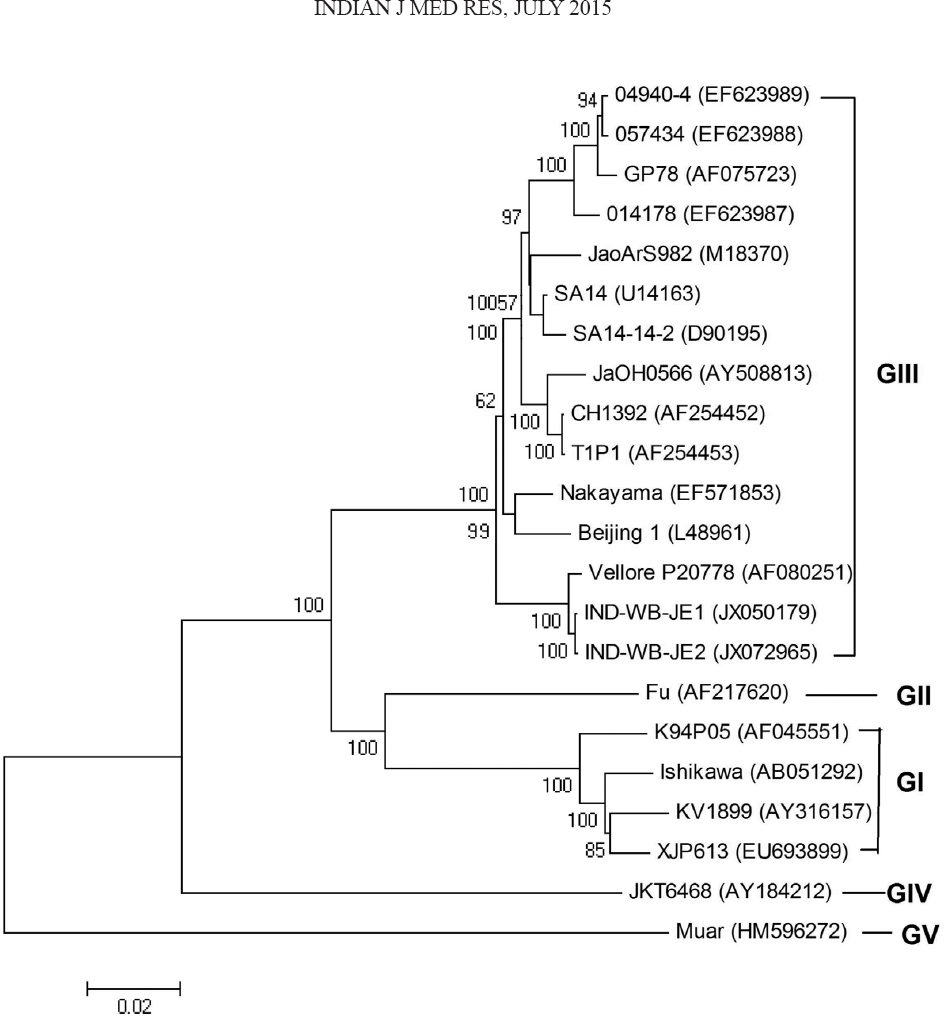
- Phylogenetic trees based on available complete nucleotide sequences of Japanese encephalitis virus. The tree was constructed using the neighbour-joining method. Horizontal branch lengths are proportional to genetic distance. Genotypes are indicated on the right. The IND-WB-JE1 and IND-WB-JE2 strains were isolated in the state of West Bengal. GenBank accession numbers for viruses shown are given in Table II.
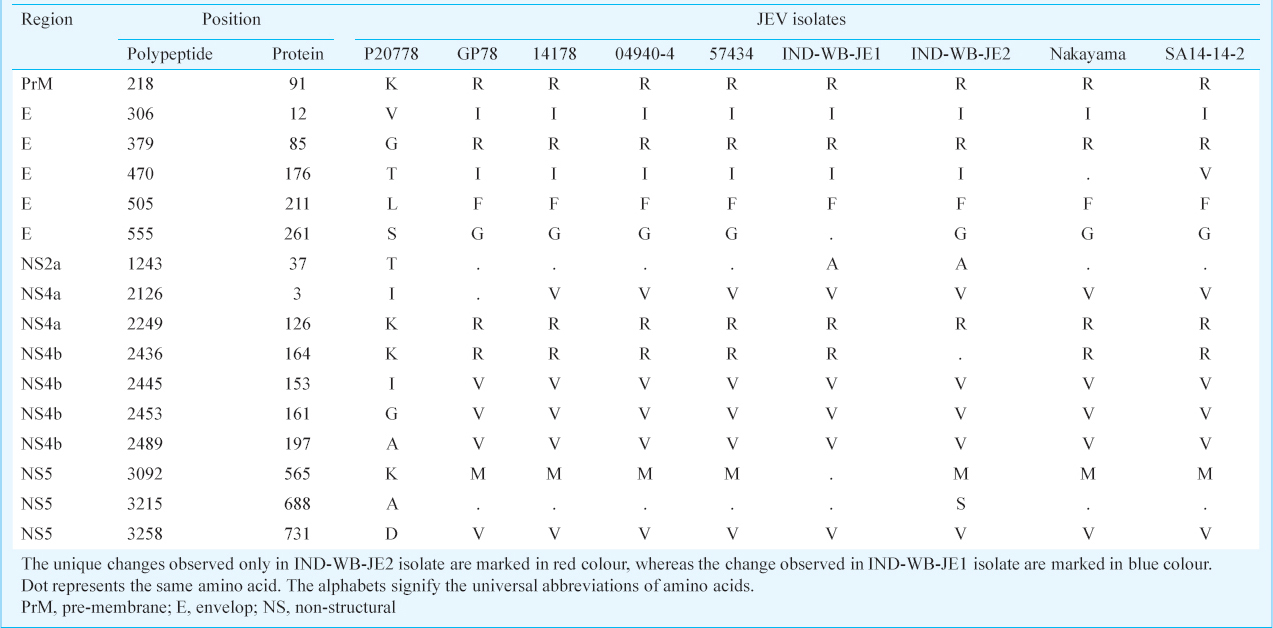
Multiple sequence alignment of amino acid sequences showed 16 non-synonymous nucleotide changes in the protein coding region of the two West Bengal isolates in comparison with the Vellore P20778 strain (Table III). In the structural protein coding region, these changes include K218R, V306I, G379R, T470I, L505F and T1243A mutations which corroborate A653G, G916A, G1135C, C1409T, G1515T and A3727G changes in the JEV genome, respectively. In non-structural protein coding region, A3727G, A6378G, A6746G, A7336G, C7469T and A9776T changes in the nucleotide corroborate I2126V, K2249R, I2445V, G2453V, A2489V and D3258V mutations in the protein coding region, respectively (Table III). Four amino acid substitutions were observed between the two West Bengal isolates. In IND-WB-JE1, K2436R (A7310G) mutation was found with respect to the Vellore-P20778 strain in NS4B gene region, but was not found in IND-WB-JE2 isolate, whereas S555G mutation (A1663G) in E protein region, K3092M (A9278T) and a unique mutation A3215S (G9645T) in the non-structural protein NS5 gene, which corroborated with A688S change in polypeptide, only found in IND-WB-JE2 isolate, but remained unchanged in the isolate IND-WB-JE1.
Discussion
Genetic variations among JEV strains isolated from widely different time periods and geographical regions have been reported in several studies. Complete genome sequences (verified) of only five isolates from human and vector mosquitoes are available in GenBank from India. We studied the full-genome sequences of two JEV isolates. The IND-WB-JE1 isolate was obtained from Malda district in 2008, where no vaccination has yet been done, and IND-WB-JE2 isolate was collected in 2010 from a 17 yr old male patient, a resident of Birbhum district, where vaccination was done in 2007.
The homology of the nucleotide and amino acid sequences was precisely compared with other JEV strains, particularly with other Indian isolates. Phylogenetically the West Bengal isolates IND-WB-JE1 and IND-WB-JE2 showed maximum homology (99% nucleotide similarity) with the first isolated strain of India i.e. Vellore P20778 (GenBank: AF080251), obtained from a patient from Vellore (south India) during 1958. This result was surprising as the two West Bengal isolates and P20778 strain were isolated from geographically distant locations at a time gap of more than 50 years. The Vellore P20778 strain was also related to Beijing 1 strain (GenBank accession no. L48961), which was isolated from a human brain in Beijing, China, in 1949. A detailed study through full genome could not be performed due to insufficient recorded data on whole genome sequences from other States of India.
From this study it became evident that JEV was still under circulation in West Bengal in both vaccine covered and not covered areas, and many deaths due to JEV infection occurred in this State even after vaccination9. This is a cause of concern and continuous monitoring on the circulating strain is required.
Acknowledgment
Authors thank Dr Sekhar Chakrabarti, the then Officer-In-Charge, ICMR Virus Unit, Kolkata, for providing facilities, and acknowledge the Department of Science and Technology, Government of West Bengal, India, for financial support.
References
- Preventive strategies for frequent outbreaks of Japanese encephalitis in Northern India. J Biosci. 2008;33:505-14.
- [Google Scholar]
- Estimated global incidence of Japanese encephalitis: a systematic review. Bull World Health Organ. 2011;89:766-74E.
- [Google Scholar]
- Molecular epidemiology of Japanese encephalitis virus, Taiwan. Emerg Infect Dis. 2010;16:876-8.
- [Google Scholar]
- Recombinant core proteins of Japanese encephalitis virus as activators of the innate immune response. Virus Genes. 2009;38:10-8.
- [Google Scholar]
- Japanese encephalitis: pathogenesis, prophylactics and therapeutics. Curr Sci. 2010;98:326-34.
- [Google Scholar]
- Virological and serological investigations of an epidemic of encephalitis which occurred at Bankura district, West Bengal. Indian J Med Res. 1976;64:121-30.
- [Google Scholar]
- Directorate of National Vector Borne Disease Control Programme, Delhi. Details of AES/JE Cases and Deaths from 2008-2013. Available from: http://nvbdcp.gov.in/Doc/ie-aes-cd-Dec13.pdf
- [Google Scholar]
- A comparison of clinical features of Japanese encephalitis virus infection in the adult and pediatric age group with acute encephalitis syndrome. J Clin Virol. 2011;52:45-9.
- [Google Scholar]
- Operational guide for Japanese encephalitis vaccination in India. New Delhi: Ministry of Health and Family Welfare, Government of India; 2010.
- [Google Scholar]
- Increasing trend of Japanese encephalitis cases in West Bengal, India - a threat to paediatric population. Asian Pac J Trop Dis. 2012;2:358-61.
- [Google Scholar]
- NCBI, nucleotide. Available from: http://www.ncbi.nlm.nih.gov/nuccore/?term=japanese+encephalitis+india++complete+genome
- [Google Scholar]
- Serological and molecular diagnosis of Japanese encephalitis reveals an increasing public health problem in the state of West Bengal, India. Trans R Soc Trop Med Hyg. 2012;106:15-9.
- [Google Scholar]
- MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596-9.
- [Google Scholar]
- Phylogenetic analysis of Japanese Encephalitis Virus: Envelope gene based analysis reveals a fifth genotype, geographic clustering, and multiple introductions of the virus in to the Indian subcontinent. Am J Trop Med Hyg. 2001;65:242-51.
- [Google Scholar]






