Circulating tumour DNA as a promising biomarker for breast cancer diagnosis & treatment monitoring
For correspondence: Dr Sudeep Gupta, Tata Memorial Centre, Homi Bhabha National Institute, Mumbai 400 012, Maharashtra, India e-mail: sudeepgupta04@yahoo.com
-
Received: ,
Accepted: ,
Abstract
Breast cancer contributes a large fraction to mortality among women diagnosed with cancer. It is important to monitor residual disease and extend the lead time to detect relapse in high-risk patients. Minimally invasive techniques that utilise circulating biomarkers are being explored for their potential in diagnosis, prognosis, and disease monitoring of breast cancer. Circulating biomarkers have been investigated as tools for breast cancer diagnosis, prognosis, prediction, and monitoring of therapeutic response and resistance. Among these, circulating tumour cells and cell-free plasma DNA (cfDNA) derived from tumour cells (circulating tumour DNA i.e. ctDNA) have been integrated into clinical trial designs. Among all circulating biomarkers, ctDNA stands out as a promising biomaterial with great potential as it is thought to mirror the tumour’s evolution. However, its clinical utilisation is hampered mainly by gaps in knowledge of its biological properties and specific characteristics. The development of robust and standardised methods for assessing circulating biomarkers is essential for realising the potential of personalised medicine. This review aims to summarise the characteristics of ctDNA and its role in breast cancer, drawing from both basic and translational research to provide insights into its clinical application. This review suggests that ctDNA has the potential to be a non-invasive, real-time surrogate for tumour tissue-based biomarkers. In conclusion, circulating biomarkers have the potential to revolutionise breast cancer diagnosis, prognosis, and treatment monitoring, but the development of standardised methods for their assessment is essential. ctDNA, in particular, shows great promise as a liquid biopsy tool, but further research is needed to understand its biology and ensure its clinical utility fully.
Keywords
Biomarkers
breast cancer
cell free DNA
circulating tumour DNA
diagnosis
liquid biopsy
Site-specific tumour biopsies are analysed to determine the best treatment for various cancers. However, the data obtained from a biopsy are transient, and the initial tumour cannot be used to examine mutations acquired during treatment or resistance development. Studies have identified up to 25 per cent of subtype mutations during or post-disease progression after anticancer treatment1. While tumour biopsy remains the gold standard for diagnosis, classifying patients, and treatment selection, liquid biopsy i.e., detection and monitoring of the cancer genome in blood samples- is advancing precision medicine. Circulating tumour cells/DNA (CTCs/ctDNA) and exosomes found in processed liquid biopsies elucidate tumour development, resistance, and heterogeneity during therapy without invasive collection procedures or sampling bias1. Thus, liquid biopsies would be the only way to accurately diagnose the tumour and understand its biology in the context of tumour heterogeneity and metastatic disease2.
The discovery of higher cell-free DNA (cfDNA) content in the peripheral blood of tumour-harbouring persons generated multiple lines of investigation into its causes and nature. Studies in the 1990s found oncogenic mutations in cfDNA sequences identical to those in tumours of the same patients, cementing the notion of its origin in tumour cells and giving rise to the term circulating tumour DNA (ctDNA), which is a type of cfDNA3. In an early study on metastatic colorectal cancer, Bardelli’s team at the University of Turin found plasma cfDNA profiling more effective than paired tissue biopsies in detecting RAS mutations4. Comparing biopsy samples with plasma samples in metastatic breast cancer (MBC) revealed that ctDNA could reflect somatic changes that represent the tumour burden and sub-clonal activity. ctDNA analysis of mutations also tracks the heterogeneity of treatment responses by various metastases and replicates the clonal hierarchy identified by multiregional cancer sequencing5. Fundamental questions of ctDNA release mechanisms and the correlation of its blood content with tumour/metastasis burden are still unanswered --despite attractive hypotheses proposed. On the translational side, attempts to harness ctDNA for clinical applications have advanced our understanding of unique features of ctDNA that enable its identification from the cfDNA pool and the correlation of these features with the prognosis and treatment response of the patient.
This article summarises studies demonstrating the use and potential of ctDNA to diagnose breast cancer, monitor therapy response, and detect minimal residual disease (MRD) or metastatic relapse post-curative treatment. Recent advances in ctDNA detection-related sensitivity and selectivity have made recognition possible, even in early-stage cancers. For instance, ctDNA levels were reported to be elevated in patients in the early stages of breast cancer when precancerous lesions were used as controls. These levels tend to increase with more advanced disease stages6,7. The clearance of ctDNA correlates with higher rates of complete pathological response after neoadjuvant treatment (NACT) and a reduced risk of recurrence following radical treatments in these patients1,8. Clinically, ctDNA is used to monitor tumour responses in real-time, identify drug-resistant clones, track changes in the tumour’s mutational profile, pinpoint actionable mutations, detect minimal residual disease, and screen for early-stage breast cancers9. While a universally accepted baseline for ctDNA in breast cancer diagnosis is still lacking, fluctuations in ctDNA levels over time can provide valuable insights into disease burden, prognosis, and therapeutic response10,11. Unlike conventional biomarkers such as Cancer Antigen 15-3 (CA 15-3) and circulating tumour cells (CTCs), ctDNA exhibits a broader dynamic range and correlates better with shifts in the tumour burden. Moreover, up to 53 per cent of patients show detectable ctDNA level changes within a month of treatment, allowing timely monitoring. Up to 53 per cent of patients show detectable ctDNA changes within a month of treatment, enabling timely monitoring12. ctDNA can also predict the outcome of the treatment. A meta-analysis by Dickinson et al13 showed that overall survival (OS) is inversely related to the absence of detectable ctDNA levels with a hazard ratio of 1.4. The study also identified significant associations of TP53 and ESR1 mutations in breast cancer patients with worse survival, while PIK3CA alterations were not associated with survival outcomes13.
The origin of ctDNA in cancer patients
The term ‘circulating free DNA’ (cfDNA) refers to small fragmented or degraded DNA pieces found in peripheral blood and range in size from 180 base pairs to 1000 base pairs. It includes cell-free foetal (cffDNA), circulating tumour (ctDNA), donor-derived cell-free (dd-cfDNA), and cell-free mitochondrial (cf-mtDNA) types of DNA14. ctDNA is the fraction of the cell-free DNA pool sourced from neoplastic cells. It makes up for up to one per cent of the total cfDNA.
Characterising ctDNA and differentiating it from cfDNA in the pre-sequencing era was done by fragment length analysis. These investigations also supported the role of apoptosis as a possible ctDNA release mechanism, as studies revealed the prevalent length of cfDNA fragments to be uniformly more than the DNA length required to wrap around a nucleosome. A nucleosome, the fundamental unit of DNA packaging in eukaryotes, comprises 144-147 bases of DNA wrapped around an octameric complex of histone proteins acting as a spool. Adjacent nucleosome cores are joined by linker DNA, which can associate with linker histone proteins such as H1 or H5. The chromatosome refers to the nucleosome core particle with approximately 165 base pairs of DNA and linker histones. cfDNA of this length provided evidence for apoptosis as a DNA release mechanism.
It was in 1977 that Leon and colleagues found that patients with cancer had more cfDNA (50 bp to 700 bp) than healthy people15. Research has demonstrated that mutations in oncogenes and/or tumour suppressors, microsatellite instability, and epigenetic marks can be revealed in ctDNA. Although ctDNA fragment profiles have been observed to differ between histologic tumour types, tumour burden, or stages of cancer, they often have lengths lower than other somatic DNA fragments found in plasma. For instance, in breast cancer, people with metastasis are likely to have shorter fragments of ctDNA than early-stage patients. While apoptotic tumours generate 180-1000 bp fragments, those from necrotic malignancies can reach 10,000 bp16,17. The size of ctDNA is roughly 20-40 bp smaller than DNA length in one nucleosome, with enrichment occurring in segments 90-150 bp and 250-320 bp in size, which also revealed that mutated ctDNA has shorter fragments than non-mutant cfDNA15,17,18. Potential reasons for this shortening include:
-
(i)
Differences present in nucleosomal wrapping or the methods of nuclease activity across varying tissue types,
-
(ii)
Variations in ctDNA size recovery due to varying methods for ctDNA extraction or
-
(iii)
Biases introduced by common techniques for extracting single-strand DNA library preps.
Processes that give rise to cfDNA
Active cellular secretion, apoptosis, and necrosis are three possible theories about cfDNA origin. cfDNA typically shows a ‘ladder’ pattern resembling apoptotic fragmentation. Depending on the nuclease activity, larger fragments such as di- tri- or poly-nuclease can also be formed due to apoptosis. Cell necrosis or programmed cell death results in the discharge of cfDNA into the bloodstream. Tissues with a high metabolic rate, such as haematopoietic and tumour tissue, tend to release more DNA. Although macrophages engulf and digest most released nucleosomes, tumour development and accelerated cell death can overload or impede this clearance system. Cancer patients in advanced stages with a higher proportion of necrotic tumour cells have more plasma ctDNA than those in early stages19,20. ctDNA could also come from CTCs and active secretion from cellular structures21. Additionally, while a tumour is made up of cancerous cells, it also includes a variety of cells that form the tumour microenvironment. Therefore, stromal cells, endothelial cells, lymphocytes, and other immune cells are all potential contributors to cfDNA release22. It has been established that the genetic alterations in ctDNA and CTC are identical. When CTCs enter the bloodstream, they encounter various physical challenges, such as the forces from blood flow, differential speed shear, interactions/impacts with other blood components, and the potential for forming complexes with non-cancerous cells like white blood cells and platelets. These factors may lead to the destruction of CTCs and the release of nucleic acids23. These results lend credence to the idea that CTC could be another source of ctDNA. However, as there are few CTCs in peripheral blood, these cells might not be the primary source of ctDNA24. According to Crowley et al25, the number of CTCs would need to be more than 2000-fold the average levels for them to account for all the ctDNA content in blood. As the occurrence of genome equivalents of ctDNA found in blood is at least a hundred times that found in CTCs, this source (if present) is of questionable relevance20. Moreover, ctDNA can often be detected even when CTCs are absent, but not the other way round26.
Having discussed the pathophysiology of ctDNA occurrence, the subsequent section summarises the potential utility of ctDNA evaluation in diagnostics, disease monitoring, drug resistance, and prognostication in breast cancer.
Uses of ctDNA
(A) Diagnostics
ctDNA in real-time can detect cancer in asymptomatic patients, monitor disease progression, and evaluate treatment response in a non-invasive manner. In some cases, ctDNA can identify tumour mutations absent in tissue biopsy, providing a comprehensive genomic profile of the tumour27. ctDNA detection strongly correlates with molecular subtypes, with the basal-like subtype exhibiting the highest ctDNA detection rate (86%), while luminal-A shows the lowest rate (0%)28. Catarino et al29 reported higher ctDNA levels in breast cancer patients than in healthy individuals (105.2 vs. 77.06 ng/ml). Furthermore, they demonstrated a notable reduction in ctDNA levels following surgery (59.0 vs. 105.2 ng/ml)29. In another study, Zhou et al30 reported that tumour-derived mutations were present in 85.71 per cent of patients with stage IV/M1 breast cancer, whereas only 57.81 per cent of stage I-III/M0 patients exhibited such mutations.
Although ctDNA is an effective biomarker in metastatic settings, ctDNA research for early breast cancer detection has yielded mixed results, as demonstrated by Kruspe et al31, showing uncertainties due to low CTC levels in early-stage cancer. Additionally, mutation rates and their specificity in ctDNA are relatively low, limiting its use in breast cancer screening. However, several instances exist where identification and analysis of specific gene mutations in cfDNA is used for breast cancer diagnosis. For instance, Arthrobacter luteus (ALU) sites that are widespread in the genome can be used to measure DNA integrity and are frequently utilised in breast cancer32. Using a fraction of ALU247 of total cfDNA levels instead of cfDNA alone led to a perfect 1.0 ROC score in differentiating between cancer-ridden and normal subjects; classifying patients with lymph node spread and those without revealed significant differences33. Another study34 correlates nodal spread using polymorphisms in specific regions of chromosome 13 and the PTEN gene using cfDNA. Methylation patterns in cfDNA have been successfully utilised for early detection using gene panels by several groups35-37. Another notable study integrated ctDNA variant calling with plasma proteins profiling in the same blood sample, achieving >99 per cent specificity38.
Detecting homologous repair deficient (HRD) tumours using BRCA variants obtained in cfDNA is a current frontier in diagnosis. Because cfDNA findings can give false negatives, no clinical guidelines specify liquid biopsy BRCA testing in lieu of conventional tests on FFPE sections39. Secondly, the official FDA label of the PARP inhibitor olaparib has no statements indicating its application in breast cancer patients with somatic BRCA mutations, and ctDNA mutations cannot be confidently identified as somatic or germline40,41. However, emerging reports demonstrate the occurrence of BRCA mutations in cfDNA of patients with HRD and compare somatic vs. germline BRCA mutation response to PARPi therapy, thus using liquid biopsies as a potential companion diagnostic42-46. Additionally, epigenetic marks like promoter methylation of BRCA1, found by Harvey-Jones et al47 in all repeat liquid biopsy samples taken from HRD tumours, demonstrate prognostic significance while circumventing the low sensitivity of ctDNA mutations.
ctDNA sequencing for cancer screening and early diagnosis remains challenging. Asymptomatic people only have ctDNA concentrations of 1 to 10 ng/ml3.Therefore, it was demonstrated that 150 to 300 ml of blood would be required for each test to reach 95 per cent sensitivity7. Second, healthy normal and haematogenic cells also supply the cfDNA pool, reducing specificity. The sensitivity and specificity standards for cancer screening and early diagnosis require considerable efforts and investments48,49. Allele-specific real-time quantitative PCR (qPCR) techniques were primarily used in the early detection of ctDNA. The sensitivity/specificity limitations made for seldom application of technologies like Peptide Nucleic Acid (PNA) clamps, probe qPCR utilising TaqMan, and Scorpion Amplification Refractory Mutation System, in patients with significant tumour burden. However, this has changed over the last decade, with techniques like digital PCR (dPCR) and next-generation sequencing (NGS) having ultrasensitive detection thresholds for mutant allele abundance between 0.01 and 0.1 per cent.
The NGS edge in ctDNA diagnostics: Though sensitive and affordable, PCR-based techniques have low input and processing speed and can only screen for known variants. However, high throughput NGS can screen unidentified variations with a detection capability of mutant allele frequency (MAF) levels below one per cent. Other techniques, such as distinct molecular IDs or barcodes, can also improve sensitivity. With a MAF of 0.1 per cent, these techniques have identified 59 per cent of patients with stage I or stage II lung cancer with good concordance between the ctDNA response and radiographic response49. Panel-based NGS can detect known variants in four ways: single locus, multiplex, targeted sequencing, and genome-wide sequencing. Several (including us) groups have recently developed NGS-based approaches that enable ultrasensitive ctDNA detection. Two of them used deep sequencing on a small set of amplicons that targeted frequently altered cancer genes. Although enabling low detection thresholds, the number of probable genomic locations has, to date, been constrained by technical issues with PCR assay multiplexing. Since most tumours won’t have a mutation identified by a specific tiny mix of amplicons, therapeutic applications are limited. The same bottleneck holds for translocations/rearrangements whose exact breakpoints aren’t known a priori50.
Custom tumour-guided sequencing panel scans cover a range of variants. Some have examined up to 115 patient-specific mutations concurrently, measuring ctDNA to one mutant molecule every 33,333 copies in breast cancer patients after NACT. Such assays typically target 10-20 variants in plasma. Current methods can detect relapse in several cancer types, such as colorectal, breast, and bladder cancer, 3-10 months sooner than clinical relapse51. As whole-genome tumour sequencing is performed more often in clinical settings, there are fewer financial and logistical obstacles to implementing patient-specific panels52.
Of all sequencing methods, multigene targeted NGS panel is most suited for ctDNA sequencing of patients lacking tumour tissue, as it enables the identification of multiple variants in a one run of analysis. However, the presence of DNA non-shedding neoplastic lesions, artefacts produced during CHIP, or sequencing distort the sensitivity and specificity of NGS-based ctDNA testing53. More research before clinical adoption is still required in this field.
(B) Minimal residual disease (MRD)
MRD (measurable, minimal or molecular residual disease) refers to residual tumour cells or biomarkers in the body post-treatment of cancer (both local and/or systemic). MRD might not exhibit any noticeable signs or symptoms and could be imperceptible with standard methods. Commonly employed techniques for MRD detection include qPCR, ddPCR, and NGS. Recent advancements in ctDNA analysis for identifying MRD in solid tumours post-curative therapy and before clinical or radiographic disease relapse have shown considerable therapeutic potential, as the detection of MRD through ctDNA analysis correlates with poor prognosis.
PCR is one of the basic techniques used to detect MRD, while BEAMing (beads, emulsion, amplification and magnetics) is a novel addition to the list. In a study by Beaver et al54, ddPCR detected PIK3CA mutations in DNA from 30 breast cancer tumours and paired plasma samples before and after surgery. Liquid biopsy accurately identified MRD, finding 14 of the 15 PIK3CA mutations in pre-surgery ctDNA and detecting the same in about 50 per cent of patients post-surgery54. However, both ddPCR and BEAMing require prior knowledge of specific mutations for detection, as they are designed to target predefined genetic changes. This restricts their ability to identify novel or unknown alterations, posing difficulties in cancers with significant heterogeneity or mutations that evolve during treatment55,56. ddPCR has a MAF threshold of about 0.1 per cent, and BEAMing has one of 0.01 per cent, but rare mutations or heavily fragmented ctDNA are often missed57,58. Additionally, they have limited multiplexing capability, a drawback when monitoring the clonal evolution of cancers such as leukaemia or lung cancer59, making it time-consuming and technically demanding for routine clinical use56,57. While ddPCR is relatively affordable, BEAMing requires specialised equipment and materials, limiting its adoption in resource-constrained settings55. In contrast, NGS, especially targeted approaches like tagged-amplicons deep sequencing (TAm-Seq) and Safe Sequencing System (Safe-SeqS), overcomes some of these limitations by enabling broader and scalable mutation detection. However, NGS lacks sensitivity compared to ddPCR and BEAMing for detecting extremely low-frequency mutations, but a combined approach could offer a more robust solution for MRD detection55,57,58.
Garcia-Murillas et al60 demonstrated that targeted capture sequencing of the ctDNA can identify MRD-related genetic events, while primary tumour sequencing was less accurate at predicting future metastatic relapses. Therefore, mutation tracking can distinguish high versus low risks of relapse in newly diagnosed breast cancer patients. Issues of tumour heterogeneity could be partially overcome by subsequent adjuvant treatment therapies that are tailored to the genetic events occurring in the MRD. Janni et al61 used an NGS panel that included about 500 genes and around 4 Mb of epigenomic areas that experience differential methylation in several solid tumour types to demonstrate the viability of MRD detection using the Guardant Reveal assay.
Recently, McDonald et al62 developed TARgeted DIgital Sequencing (TARDIS), which uses exome sequencing of tumour biopsies to identify somatic mutations. Targeted linear pre-amplification of ctDNA with specific primers increases sensitivity. The test accurately detected ctDNA in all 33 patients tested--and also detected trace amounts in 17 out of 22 patients receiving NACT. Preliminary results in early-stage breast cancer suggest TARDIS can detect MRD with over 100 times the sensitivity of current methods62. Various studies that have demonstrated ctDNA as an independent indicator of the residual disease are described in table I8,50,63,64.
| Sr. No. | ctDNA concentration | Study outcome | Reference | |
|---|---|---|---|---|
| Baseline time-point | Progression time-point | |||
| 1 | 1 copy per ml (0.2-8.9) in non-metastatic patients | 3.1 copies per ml median (0.1-1145.6) in metastatic patients (9 months post baseline) | 5/32 patients in the intervention group with ctDNA+ but no upfront metastasis, relapsed with concurrent failure of ctDNA clearance. | 63 |
| 2 | 14.825 ng median yield (1.49-72.38) at T0 time-point | 33.965 ng median yield (12.64-267.07) at T2-T3 time-point | A positive correlation of ctDNA presence at timepoint T1 (83%; 24/29 vs 52%; 14/27) with residual tumour after surgery was found | 8 |
| 3 | 13.5 mean ctDNA yield at baseline | 17 mean ctDNA yield at fourth time-point | Area under curve (AUC) values for ROC curve for 9 TNBC patients and 11 ER+/HER2± patients found to be 1 and 0.89, respectively when predicting residual tumour using clearance status of ctDNA post neo-adjuvants | 50 |
| 4 | cfDNA yield 30.7 ng (no. of patients = 74) | cfDNA yield 38.5ng (no. of patients = 63) | Mutant allele fraction followed patient response to therapy versus progression. cfDNA yield/circulating tumour cells only rose with the progression | 64 |
(C) Therapeutic resistance
ctDNA is increasingly used to detect therapeutic resistance in cancer treatment. As tumours evolve under treatment pressure, they may acquire genetic mutations that confer resistance to therapies, such as targeted treatments or chemotherapy.
Chen et al65 identified resistance-correlated genes in liquid biopsies using NGS. Activating mutations in PIK3CA (C420R), TP53 and in HER2 (L869R) had correlations with HER2 targeted therapy resistance. Thompson et al66 highlight the advantages of using ctDNA NGS to detect driver and resistance mutations, even when tissue samples are unavailable. While ctDNA testing alone cannot identify histologic sources of resistance, such as a small-cell phenotype, it can offer more detailed molecular characterisation of the tumour. The study also suggests that higher levels of cfDNA are linked to poorer survival outcomes, regardless of the mutational profile.
Unlike traditional imaging and serum biomarkers like CEA and CA15-3, which lack sensitivity for detecting minor tumour mutations, ctDNA provides a prompt and sensitive alternative for monitoring cancer progression. Specific ctDNA mutations can also guide treatment decisions, with examples including PIK3CA and BRCA1 mutations in MBC that suggest sensitivity to drugs like everolimus, alpelisib, and Olaparib, respectively. How ctDNA mutation patterns relate to hormone receptor/HER2 status and therapy is still unclear. A study by Davidson et al67 showed that targeting AKT1 and ERBB2 mutations in ctDNA with capivasertib and neratinib, respectively, led to partial responses in 4/18 and 5/20 patients, suggesting the potential of mutation-driven therapies in late-stage, pre-treated breast cancer64. Jacob et al68 demonstrated that when comparing with tissue NGS, alterations detected in ctDNA at baseline showed a high degree of concordance. Notably, most of the tissue samples were taken from sites of metastatic disease, suggesting that resistance alterations may arise earlier in disease progression. Another explanation is that modest to moderate increases in clonal heterogeneity may not be captured due to dominant clones found in tissue samples.
A landmark study by Mosele et al69 demonstrated the prevalence and clinical implications of PIK3CA mutations in different breast cancer subtypes. Among HR+/HER2- tumours, 28 per cent harboured a PIK3CA mutation, compared to 10 per cent in triple-negative breast cancer (TNBC). PIK3CA-mutated HR+/HER2- MBC showed reduced chemotherapy sensitivity and worse OS, whereas, in metastatic TNBC, PIK3CA mutation was associated with improved median OS66. Continuously tracking treatment responses and ctDNA mutations in patients with MBC, there is potential to tackle drug resistance, though this requires confirmation through extensive prospective trials60. In summary, ctDNA holds significant promise in oncology, particularly in advancing personalised medicine.
(D) CtDNA as a prognostic marker for outcomes and response
Along with monitoring MRD and treatment resistance, ctDNA is also explored as a prognostic marker for survival and outcomes. Pècuchet et al70 determined instead of being interpreted for its baseline concentration, the measurement of ctDNA in plasma throughout treatment should be evaluated independently. While baseline ctDNA measurements can provide initial insights into tumour burden and prognosis, they cannot capture tumour heterogeneity and predict treatment resistance. However, by serially measuring ctDNA levels over time, clinicians can detect early signs of recurrence, assess treatment response, identify emerging resistance mechanisms, and tailor treatment strategies71,72. For instance, patients with initially low baseline ctDNA levels but a rapid increase during treatment had significantly worse outcomes compared to those with stable or declining levels73. Thus, serial monitoring can identify high-risk patients who may need aggressive therapeutic interventions.
The absence of ctDNA normalisation in the initial assessment has a strong prognostic impact on both progression-free survival (PFS) and OS. As opposed to the MAF, which is a relative percentage statistic, the ctDNA concentration reported in studies is an absolute value of the MAF. Studies detected an inverse correlation between baseline ctDNA amount and the total amount of cfDNA and OS74,75. Poorer outcomes are associated with ctDNA levels in early, locally advanced, and metastatic diseases.
Quality control for ctDNA
Blood is collected in BCTs, such as EDTA, Paxgene, and Strecktubes having proprietary elements that preserve the cfDNA and prevent cell lysis. Plasma samples are considered a better source of ctDNA as they provide wild-type DNA fractions. The overall cfDNA concentration in serum is 2-24 times higher, which is likely due to the contamination from DNA released from immune cells. Since clotting induces lysing of these immune cells, less contamination is found in plasma76. Firstly, for extraction of cfDNA, blood is centrifuged at 1200×g for 10 min, then again at 16000×g for 10 min at 4°C. An overview of extraction and analysis of ctDNA from the resulting plasma layer has been illustrated in figure.
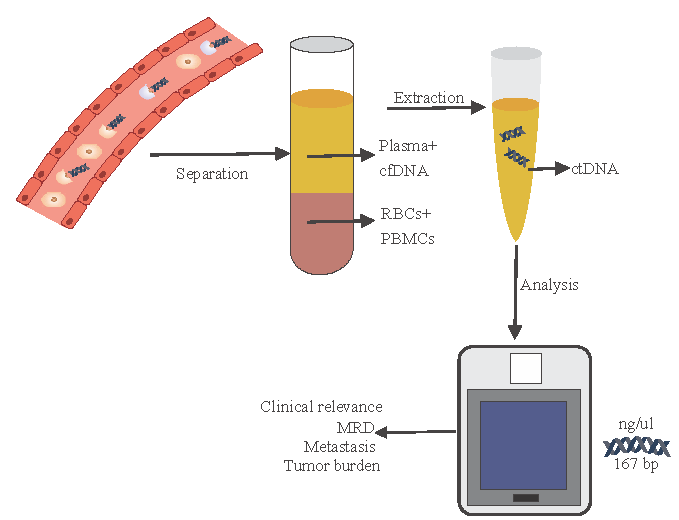
- Extraction and analysis pipeline for circulating tumour DNA. The figure was created using CorelDraw 2024 (https://www.coreldraw.com/).
Available kits for cfDNA extraction kits employ differing principles: (a) column-based separation, (b) spin column chromatography, and (c) size-based separation. Among these kits, column-based kits demonstrated better performance compared to the other two. This kit was the most effective, with a better representation of tiny-fragmented DNA. It also demonstrated maximum recovery of cfDNA and permitted an effective recovery of a spike-in. The basic column-based DNA extraction from whole blood kits was found to be more comparable to the column-based extraction kits by Repiská et al77. However, a possible confounding effect is introduced by different adaptations of the DNA from whole blood kits for cfDNA extraction. An overview comparison of prevalent extraction methods is given in table II78-80.
| Method | Strengths | Weaknesses |
|---|---|---|
| Silica Membrane | High yield, established protocols | Time-consuming, risk of contamination |
| Magnetic Bead | Fast, easy automation | Potentially lower yields in some cases |
| Phenol-Chloroform | High-quality extraction | Toxic reagents, labour-intensive |
| Filtration | Quick processing | Variable yields, less commonly used |
| Liquid-Phase (ATPS) | Higher yields, better mutant recovery | Less established, still under evaluation |
Extracted cfDNA is quantified using various kits and instruments as follows: (a) Qubit, (b) Tapestation, (c) Bioanalyzer, and (d) PCR. Fluorometric quantification, a streamlined approach compared to qPCR, has gained prominence in clinical laboratories due to its efficiency. By employing fluorescent probes selectively binding double-stranded DNA, fluorometry addresses the limitations inherent in spectrophotometry-based methods, such as NanoDrop. For downstream analyses requiring both quantification and cfDNA sizing, the concurrent utilisation of electrophoresis-based techniques, exemplified by TapeStation with various assays, is strongly recommended. Acceptable quality control value ranges for extracted ctDNA are described in table III81-83. Furthermore, electrophoresis-based methods can be instrumental in assessing the quality of isolated cfDNA, particularly when pre-analytical factors, such as sample storage and shipping, may introduce the risk of gDNA contamination from cell lysis81-83.
| QC Parameter | Qubit | Tapestation |
|---|---|---|
| Size | 500 bp | 50-700 bp |
| Integrity | NA | >2.6 |
| Concentration | >30 ng/ul | 100-4000 pg/ul |
In summary, we note how ctDNA analysis complements conventional biopsy to provide a more representative sample of tumour mutations in a non-invasive manner. Its correlation with tumour burden makes it ideal for monitoring disease progress, especially in the MRD setting. The main innovations required before ctDNA tests become a routine diagnostic tool include better methods of identifying the ctDNA population in the liquid biopsy, research into novel prognostic and actionable data informed by ctDNA analysis specific to cancer subtypes and lowering costs of sequencing. As with all novel techniques, awareness of the transforming impacts of ctDNA testing among clinicians remains the primary factor deciding its adoption in routine practice.
Financial support & sponsorship
None.
Conflicts of Interest
None.
Use of Artificial Intelligence (AI)-Assisted Technology for manuscript preparation
The authors confirm that there was no use of AI-assisted technology for assisting in the writing of the manuscript and no images were manipulated using AI.
References
- Role of ctDNA in breast cancer. Cancers (Basel). 2022;14:310.
- [CrossRef] [PubMed] [PubMed Central] [Google Scholar]
- Circulating tumour DNA assays in clinical cancer research. J Natl Cancer Inst. 2018;110:929-34.
- [CrossRef] [PubMed] [PubMed Central] [Google Scholar]
- Liquid biopsies come of age: Towards implementation of circulating tumour DNA. Nat Rev Cancer. 2017;17:223-38.
- [CrossRef] [PubMed] [Google Scholar]
- Dynamic molecular analysis and clinical correlates of tumour evolution within a phase II trial of panitumumab-based therapy in metastatic colorectal cancer. Ann Oncol. 2018;29:119-26.
- [CrossRef] [PubMed] [PubMed Central] [Google Scholar]
- Multifocal clonal evolution characterized using circulating tumour DNA in a case of metastatic breast cancer. Nat Commun. 2015;6:8760.
- [CrossRef] [PubMed] [PubMed Central] [Google Scholar]
- Measurement of circulating cell-free DNA levels by a simple fluorescent test in patients with breast cancer. Am J Clin Pathol. 2015;143:18-24.
- [CrossRef] [PubMed] [Google Scholar]
- The plasma DNA concentration as a potential breast cancer screening marker. Indian J Clin Biochem. 2015;30:55-8.
- [CrossRef] [PubMed] [PubMed Central] [Google Scholar]
- Circulating tumour DNA in neoadjuvant-treated breast cancer reflects response and survival. Ann Oncol. 2021;32:229-39.
- [CrossRef] [PubMed] [PubMed Central] [Google Scholar]
- Circulating tumour cells and circulating tumour DNA in breast cancer diagnosis and monitoring. Oncol Res. 2023;31:667-75.
- [CrossRef] [PubMed] [PubMed Central] [Google Scholar]
- Cell-free circulating tumour DNA analysis for breast cancer and its clinical utilization as a biomarker. Oncotarget. 2017;8:75742-55.
- [CrossRef] [PubMed] [PubMed Central] [Google Scholar]
- The dynamic range of circulating tumour DNA in metastatic breast cancer. Breast Cancer Res. 2014;16:421.
- [CrossRef] [PubMed] [PubMed Central] [Google Scholar]
- Analysis of circulating tumour DNA to monitor metastatic breast cancer. N Engl J Med. 2013;368:1199-209.
- [CrossRef] [PubMed] [Google Scholar]
- Circulating tumour DNA and survival in metastatic breast cancer: A systematic review and meta-analysis. JAMA Netw Open. 2024;7:e2431722.
- [CrossRef] [PubMed] [PubMed Central] [Google Scholar]
- Life and death of circulating cell-free DNA. Cancer Biol Ther. 2019;20:1057-67.
- [CrossRef] [PubMed] [PubMed Central] [Google Scholar]
- Free DNA in the serum of cancer patients and the effect of therapy. Cancer Res. 1977;37:646-50.
- [PubMed] [Google Scholar]
- Enhanced detection of circulating tumour DNA by fragment size analysis. Sci Transl Med. 2018;10:eaat4921.
- [CrossRef] [PubMed] [PubMed Central] [Google Scholar]
- Circulating tumour-derived DNA is shorter than somatic DNA in plasma. Proc Natl Acad Sci US. 2015;112:3178-9.
- [Google Scholar]
- Increased detection of circulating tumour DNA by short fragment enrichment. Transl Lung Cancer Res. 2021;10:1501-11.
- [CrossRef] [PubMed] [PubMed Central] [Google Scholar]
- Clinical relevance of circulating nucleosomes in cancer. Ann N Y Acad Sci. 2008;1137:180-9.
- [CrossRef] [PubMed] [Google Scholar]
- Origins, structures, and functions of circulating DNA in oncology. Cancer Metastasis Rev. 2016;35:347-76.
- [CrossRef] [PubMed] [PubMed Central] [Google Scholar]
- The importance of examining the proportion of circulating DNA originating from tumour, microenvironment and normal cells in colorectal cancer patients. Expert Opin Biol Ther. 2012;12:S209-15.
- [CrossRef] [PubMed] [Google Scholar]
- Circulating tumour nucleic acids: biology, release mechanisms, and clinical relevance. Mol Cancer. 2023;22:15.
- [CrossRef] [PubMed] [PubMed Central] [Google Scholar]
- Circulating tumour DNA: a promising biomarker in the liquid biopsy of cancer. Oncotarget. 2016;7:48832-41.
- [CrossRef] [PubMed] [PubMed Central] [Google Scholar]
- Liquid biopsy: monitoring cancer-genetics in the blood. Nat Rev Clin Oncol. 2013;10:472-84.
- [CrossRef] [PubMed] [Google Scholar]
- Detection of circulating tumour DNA in early- and late-stage human malignancies. Sci Transl Med. 2014;6:224ra24.
- [CrossRef] [PubMed] [PubMed Central] [Google Scholar]
- Natural history of germline BRCA1 mutated and BRCA wild-type triple-negative breast cancer. Cancer Res Commun. 2024;4:404.
- [CrossRef] [PubMed] [PubMed Central] [Google Scholar]
- Circulating tumour DNA in HER2-amplified breast cancer: A translational research substudy of the NeoALTTO phase III trial. Clin Cancer Res. 2019;25:3581-8.
- [CrossRef] [PubMed] [Google Scholar]
- Quantification of free circulating tumour DNA as a diagnostic marker for breast cancer. DNA Cell Biol. 2008;27:415-21.
- [CrossRef] [PubMed] [Google Scholar]
- Clinical factors associated with circulating tumour DNA (ctDNA) in primary breast cancer. Mol Oncol. 2019;13:1033-46.
- [CrossRef] [PubMed] [PubMed Central] [Google Scholar]
- Rapid and sensitive detection of breast cancer cells in patient blood with nuclease-activated probe technology. Mol Ther Nucleic Acids. 2017;8:542-57.
- [CrossRef] [PubMed] [PubMed Central] [Google Scholar]
- Cell-free DNA fragmentomics: A promising biomarker for diagnosis, prognosis and prediction of response in breast cancer. Int J Mol Sci. 2022;23:14197.
- [CrossRef] [PubMed] [PubMed Central] [Google Scholar]
- Circulating cell-free DNA: A promising marker of regional lymphonode metastasis in breast cancer patients. Cancer Biomark. 2012;11:89-98.
- [CrossRef] [PubMed] [Google Scholar]
- Evaluation of cell-free tumour DNA and RNA in patients with breast cancer and benign breast disease. Mol Biosyst. 2011;7:2848-54.
- [CrossRef] [PubMed] [Google Scholar]
- Promoter hypermethylation of the tumour-suppressor genes ITIH5, DKK3, and RASSF1A as novel biomarkers for blood-based breast cancer screening. Breast Cancer Res. 2013;15:R4.
- [CrossRef] [PubMed] [PubMed Central] [Google Scholar]
- A DNA methylation-based test for breast cancer detection in circulating cell-free DNA. J Clin Med. 2018;7:420.
- [CrossRef] [PubMed] [PubMed Central] [Google Scholar]
- Cell-free DNA methylation of selected genes allows for early detection of the major cancers in women. Cancers (Basel). 2018;10:357.
- [CrossRef] [PubMed] [PubMed Central] [Google Scholar]
- Detection and localization of surgically resectable cancers with a multi-analyte blood test. Science. 2018;359:926-30.
- [CrossRef] [PubMed] [PubMed Central] [Google Scholar]
- ESMO recommendations on the use of circulating tumour DNA assays for patients with cancer: A report from the ESMO precision medicine working group. Ann Oncol. 2022;33:750-68.
- [CrossRef] [PubMed] [Google Scholar]
- FDA D.I.S.C.O.: Olaparib for gBRCAm HER2-negative metastatic breast cancer. Available from: https://www.fda.gov/drugs/resources-information-approved-drugs/fda-disco-olaparib-gbrcam-her2-negative-metastatic-breast-cancer, accessed on April 2, 2025.
- Prevalence of pathogenic germline variants in the circulating tumour DNA testing. Int J Clin Oncol. 2022;27:1554-61.
- [CrossRef] [PubMed] [PubMed Central] [Google Scholar]
- TBCRC 048: A phase II study of olaparib monotherapy in metastatic breast cancer patients with germline or somatic mutations in DNA damage response (DDR) pathway genes (Olaparib Expanded) J Clin Oncol. 2020;38:1002.
- [CrossRef] [Google Scholar]
- Circulating tumour DNA (ctDNA) as a tool to help guide germline testing in patients with solid malignancies. J Clin Oncol. 2021;39:10602.
- [Google Scholar]
- Olaparib monotherapy as primary treatment in unselected triple negative breast cancer. Ann Oncol. 2021;32:240-9.
- [CrossRef] [PubMed] [Google Scholar]
- Circulating tumour DNA validity and potential uses in metastatic breast cancer. NPJ Breast Cancer. 2024;10:21.
- [CrossRef] [PubMed] [PubMed Central] [Google Scholar]
- Olaparib: A novel therapy for metastatic breast cancer in patients with a BRCA1/2 mutation. J Adv Pract Oncol. 2019;10:167.
- [CrossRef] [PubMed] [PubMed Central] [Google Scholar]
- Abstract 6094: Longitudinal analysis of PARP inhibitor and platinum resistance in BRCA1/2m breast cancer using liquid biopsy. Cancer Res. 2023;83:6094.
- [Google Scholar]
- ctDNA monitoring using patient-specific sequencing and integration of variant reads. Sci Transl Med.. 2020;12:eaaz8084.
- [CrossRef] [PubMed] [Google Scholar]
- Next-generation sequencing in liquid biopsy: cancer screening and early detection. Hum Genomics. 2019;13:34.
- [CrossRef] [PubMed] [PubMed Central] [Google Scholar]
- Personalized circulating tumour DNA analysis to detect residual disease after neoadjuvant therapy in breast cancer. Sci Transl Med. 2019;11:eaax7392.
- [CrossRef] [PubMed] [PubMed Central] [Google Scholar]
- Serial circulating tumour DNA analysis during multimodality treatment of locally advanced rectal cancer: a prospective biomarker study. Gut. 2019;68:663-71.
- [CrossRef] [PubMed] [PubMed Central] [Google Scholar]
- Combined circulating tumour DNA and protein biomarker-based liquid biopsy for the earlier detection of pancreatic cancers. Proc Natl Acad Sci USA. 2017;114:10202-7.
- [CrossRef] [PubMed] [PubMed Central] [Google Scholar]
- Next generation sequencing-based profiling of cell free DNA in patients with advanced non-small cell lung cancer: Advantages and pitfalls. Cancers (Basel). 2020;12:1-17.
- [CrossRef] [Google Scholar]
- Detection of cancer DNA in plasma of patients with early-stage breast cancer. Clin Cancer Res. 2014;20:2643-50.
- [CrossRef] [PubMed] [PubMed Central] [Google Scholar]
- High-throughput droplet digital PCR system for absolute quantitation of DNA copy number. Anal Chem. 2011;83:8604-10.
- [CrossRef] [PubMed] [PubMed Central] [Google Scholar]
- BEAMing: single-molecule PCR on microparticles in water-in-oil emulsions. Nat Methods. 2006;3:551-9.
- [CrossRef] [PubMed] [Google Scholar]
- Circulating tumour DNA detection in MRD assessment and diagnosis and treatment of non-small cell lung cancer. Front Oncol. 2022;12:1027664.
- [CrossRef] [PubMed] [PubMed Central] [Google Scholar]
- An ultrasensitive method for quantitating circulating tumour DNA with broad patient coverage. Nat Med. 2014;20:548-54.
- [CrossRef] [PubMed] [PubMed Central] [Google Scholar]
- Comparison of BEAMing and droplet digital PCR for circulating tumour DNA analysis. Clin Chem. 2019;65:1405-13.
- [CrossRef] [PubMed] [Google Scholar]
- Mutation tracking in circulating tumour DNA predicts relapse in early breast cancer. Sci Transl Med. 2015;7:302ra133.
- [CrossRef] [PubMed] [Google Scholar]
- Abstract 3403: Multiomic, plasma-only circulating tumour DNA (ctDNA) assay identifies breast cancer patients with minimal residual disease (MRD) and predicts distant recurrence. Cancer Res. 2022;82:3403.
- [Google Scholar]
- Abstract P4-01-21: Multiplexed targeted digital sequencing of circulating tumour DNA to detect minimal residual disease in early and locally advanced breast cancer. Cancer Res. 2019;79:P4. 01-21-P4-01-21
- [Google Scholar]
- Results of the c-TRAK TN trial: a clinical trial utilising ctDNA mutation tracking to detect molecular residual disease and trigger intervention in patients with moderate- and high-risk early-stage triple-negative breast cancer. Ann Oncol. 2023;34:200-11.
- [CrossRef] [PubMed] [Google Scholar]
- Longitudinal dynamics of circulating tumour cells and circulating tumour DNA for treatment monitoring in metastatic breast cancer. JCO Precis Oncol. 2021;5:943-52.
- [CrossRef] [PubMed] [PubMed Central] [Google Scholar]
- Monitoring treatment efficacy and resistance in breast cancer patients via circulating tumour DNA genomic profiling. Mol Genet genomic Med. 2020;8:e1079.
- [CrossRef] [PubMed] [PubMed Central] [Google Scholar]
- Detection of therapeutically targetable driver and resistance mutations in lung cancer patients by next-generation sequencing of cell-free circulating tumour DNA. Clin Cancer Res. 2016;22:5772-82.
- [CrossRef] [PubMed] [PubMed Central] [Google Scholar]
- The breast is yet to come: current and future utility of circulating tumour DNA in breast cancer. Br J Cancer. 2021;125:780-8.
- [CrossRef] [PubMed] [PubMed Central] [Google Scholar]
- The use of serial circulating tumour DNA to detect resistance alterations in progressive metastatic breast cancer. Clin Cancer Res. 2021;27:1361-70.
- [CrossRef] [PubMed] [Google Scholar]
- Outcome and molecular landscape of patients with PIK3CA-mutated metastatic breast cancer. Ann Oncol. 2020;31:377-86.
- [CrossRef] [PubMed] [Google Scholar]
- Base-position error rate analysis of next-generation sequencing applied to circulating tumour DNA in non-small cell lung cancer: A prospective study. PLoS Med. 2016;13:e1002199.
- [CrossRef] [PubMed] [PubMed Central] [Google Scholar]
- Non-invasive analysis of acquired resistance to cancer therapy by sequencing of plasma DNA. Nature. 2013;497:108-12.
- [CrossRef] [PubMed] [Google Scholar]
- Liquid biopsy: a step closer to transform diagnosis, prognosis and future of cancer treatments. Mol Cancer. 2022;21:1-22.
- [CrossRef] [PubMed] [PubMed Central] [Google Scholar]
- Changes in circulating tumour DNA and outcomes in solid tumours treated with immune checkpoint inhibitors: A systematic review. J Immunother Cancer. 2023;11:e005854.
- [CrossRef] [PubMed] [PubMed Central] [Google Scholar]
- Circulating tumour DNA clearance predicts prognosis across treatment regimen in a large real-world longitudinally monitored advanced non-small cell lung cancer cohort. Transl Lung Cancer Res. 2020;9:269-79.
- [CrossRef] [PubMed] [PubMed Central] [Google Scholar]
- Emerging role of genomics and cell-free DNA in breast cancer. Curr Treat Options Oncol. 2019;20:68.
- [CrossRef] [PubMed] [Google Scholar]
- Plasma or serum: Which is preferable for mutation detection in liquid biopsy? Clin Chem. 2020;66:946-57.
- [CrossRef] [PubMed] [Google Scholar]
- Selection of the optimal manual method of cell free fetal DNA isolation from maternal plasma. Clin Chem Lab Med. 2013;51:1185-9.
- [CrossRef] [PubMed] [Google Scholar]
- Comparison of methods for circulating cell-free DNA isolation using blood from cancer patients: Impact on biomarker testing. Transl lung cancer Res. 2016;5:665-72.
- [CrossRef] [PubMed] [PubMed Central] [Google Scholar]
- Precision cell-free DNA extraction for liquid biopsy by integrated microfluidics. NPJ Precis Oncol. 2020;4:3.
- [CrossRef] [PubMed] [PubMed Central] [Google Scholar]
- Isolation and quantification of plasma cell-free DNA using different manual and automated methods. Diagnostics (Basel). 2022;12:2550.
- [CrossRef] [PubMed] [PubMed Central] [Google Scholar]
- Comprehensive evaluation of methods to isolate, quantify, and characterize circulating cell-free DNA from small volumes of plasma. Anal Bioanal Chem. 2015;407:6873-8.
- [CrossRef] [PubMed] [Google Scholar]
- Optimizing the yield and utility of circulating cell-free DNA from plasma and serum. Clin Chim Acta. 2009;404:100-4.
- [CrossRef] [PubMed] [Google Scholar]
- Towards standardisation of cell-free DNA measurement in plasma: controls for extraction efficiency, fragment size bias and quantification. Anal Bioanal Chem. 2014;406:6499-512.
- [CrossRef] [PubMed] [PubMed Central] [Google Scholar]






