Translate this page into:
Characterization of blaOXA-232 carrying carbapenem-resistant Klebsiella pneumoniae (CRKP) & their expression profiles under selective carbapenem pressure: An in-depth study from India
For correspondence: Dr Amitabha Bhattacharjee, Department of Microbiology, Assam University, Silchar, 788 011, Assam, India e-mail: ab0404@gmail.com
-
Received: ,
Abstract
Background & objectives
OXA-232 is a five amino acid substitution variant of OXA-48 and is reported in carbapenem-resistant Klebsiella pneumoniae (CRKP), which is associated with nosocomial infections among immunocompromised patients in the intensive care unit. This study aimed to characterise blaOXA-232 in CRKP of clinical origin and investigate its transcriptional response against sub-inhibitory levels of carbapenems.
Methods
CRKP was isolated from blood (pathogens) and stool cultures (colonisers) of neonates and was characterized for blaOXA-232. Co-existing resistance determinants were investigated in blaOXA-232 positive isolates, followed by horizontal gene transferability assay and PCR-based replicon typing (PBRT). Cloning of blaOXA-232 was performed, and expression of blaOXA-232 in the isolates and their clones under sub-inhibitory concentrations of carbapenems was checked via RT-PCR. Mobile genetic elements associated with blaOXA-232 were investigated, followed by DNA fingerprinting through enterobacterial repetitive intergenic consensus (ERIC) PCR.
Results
blaOXA-232 with co-carriage of extended-spectrum beta-lactamases (ESBLs), sulphonamides and quinolones were identified in seven CRPK isolates recovered from blood samples of neonates. Transformation and cloning of blaOXA-232 was successful. The sub-inhibitory concentration of carbapenems induces elevated expression of this resistant determinant. ISEcp1 was associated with blaOXA-232 in the upstream region within two haplotypes of CRKP isolates of clinical origin.
Interpretation & conclusions
Selective carbapenem pressure resulted in higher expression of this gene, which could account for treatment failure. With frequent reports of occurrence among clinical isolates, monitoring and further investigation of this novel variant are necessary to understand its transmission dynamics and to thwart its further dissemination.
Keywords
Antimicrobial resistance
CRKP
ISEcp1
neonatal septicaemia
OXA-232
Over the last two decades, Klebsiella pneumoniae (K. pneumoniae) has emerged as a clinically significant infectious agent due to its association with nosocomial infections, often causing considerable morbidity and mortality. Several cases of infections have been documented to date, particularly in patients with impaired immune systems in the intensive care unit (ICU), where this pathogen has successfully adapted itself through various mechanisms1. Treating such infections is a major challenge to clinicians worldwide because of resistance towards multiple drugs, including carbapenems, the last resort antibiotic in treating multidrug-resistant (MDR) infections2-4. Carbapenem-resistant K. pneumoniae (CRKP) has been grouped under ‘critical priority’ pathogens owing to its pathogenicity, multidrug resistance, transmission and dearth of available treatment options by the World Health Organization5. Even in the Indian scenario, CRKP has been recognized as a pathogen of great concern6. The investigators’ lab has characterised CRKP and hypervirulent K. pneumoniae (hvKP) belonging to ST5235 from cases of neonatal sepsis with high mortality occurring during multiple outbreaks of K. pneumoniae in the neonatal intensive care unit (NICU) of a tertiary referral hospital7,8.
Since their first report in 2009, the New Delhi metallo-beta-lactamase (NDM) and its variants, became the most widely reported carbapenemase and were predominant in the Indian subcontinent9. However, of late, the resurgence of reports of OXA-48-type enzymes worldwide among Carbapenem-Resistant Enterobacterales (CRE), including K. pneumoniae has posed new challenges in their management10. OXA-232, a five amino acid substitution variant of OXA-48, the class D carbapenemase responsible for carbapenem resistance was first reported within CRKP from France in 2013. It was found localized to a 6.1 kb non-conjugative ColE plasmid11,12. Since its first report, this OXA-48 variant has been detected in Singapore, USA, Italy, China and India within CRKP of clinical origin in neonates as well as adult patients suffering from severe infections10,12-18.
In comparison to OXA-48, the variant shows lower hydrolytic activity against carbapenems. Despite this, several K. pneumoniae isolates harbouring the blaOXA-232 gene have been reported to be carbapenem-resistant19. Hence, it is important to understand the behaviour of these isolates when exposed to sub-inhibitory levels of carbapenems. With recent reports of this emerging variant from India and paucity of information available about the factors triggering its adaptive response, the present study investigates the incidence of blaOXA-232 within CRKP of clinical origin and its transcriptional response against concentration-dependent carbapenem stress at sub-inhibitory levels.
Material & Methods
This was a descriptive study characterizing the CRKP isolates collected from outbreaks of neonatal sepsis, with special reference to blaOXA-232 gene. This research was carried out in the department of Microbiology, Assam University, Silchar. Thirty five non-duplicate clinical strains of K. pneumoniae exhibiting non-susceptibility towards anyone of the carbapenem were received from the Infection Control Section, department of Microbiology, Institute of Medical Sciences, Banaras Hindu University, Varanasi after obtaining approval from Institutional Ethical Committee.
Study setting and bacterial isolates
The isolates were part of the surveillance and outbreak control practices initiated in the neonatal intensive care unit (NICU) during the period 2017-2022. Briefly, during this period, three outbreaks of K. pneumoniae were reported from the NICU. As a part of the hospital infection control protocol, every outbreak was investigated with microbiological environmental surveillance, stool/anal swab culture surveillance and determination of clonality of the isolates either through phenotypic antibiogram typing or genotypically through enterobacterial repetitive intergenic consensus (ERIC) PCR. For this study, only those isolates from the blood or stool of neonates were included, which were non-susceptible to any one carbapenem (imipenem/meropenem/ertapenem). Only one or two isolates (in case of monoclonal outbreak) from each cluster were considered in the study. Inappropriately stored isolates and those without adequate clinical data were excluded. The isolates were phenotypically identified by biochemical methods and were further confirmed via VITEK®2 Compact System (Biomerieux, France).
Antibiotic susceptibility testing
Kirby-Bauer disc diffusion method was used to determine the susceptibility of the study isolates against cefepime (30 µg), ceftriaxone (30 µg), ceftazidime (30 µg), aztreonam (30 µg), imipenem (10 µg), meropenem (10 µg), ertapenem (10 µg), piperacillin/tazobactam (100/10 µg), amikacin (10 µg), gentamicin (10 µg), ciprofloxacin (5 µg) and levofloxacin (5 µg) (HiMedia Laboratories Pvt. Ltd.). The susceptibility patterns were interpreted according to the Clinical and Laboratory Standard Institute (CLSI) 2022 breakpoint criteria for Enterobacterales. Escherichia coli ATCC 25922 and Pseudomonas aeruginosa ATCC 27853 were used as controls.
Broth microdilution method (BMD) was performed as per CLSI 2022 guidelines to establish the Minimum Inhibitory Concentrations (MICs) of imipenem (Merck, France), meropenem (AstraZeneca, UK) and ertapenem (MSD, France) against the study isolates20.
Screening of carbapenemase production
Carbapenemase production among the study isolates was phenotypically investigated using Rapidec® Carba NP (Biomerieux, France) in accordance with the manufacturer’s directives. E. coli ATCC 25922 was used as a negative control for the experiment.
Molecular characterization of blaOXA-232
DNA templates for the PCR assay were extracted by the boiling centrifugation method21. The primer pair OXA-232F (5/-ATTATCGGAATGCCAGCGGT-3/) and OXA-232R (5/-AGGGCGATCAAGCTATTGGG-3/) designed using the NCBI primer blast tool (https://www.ncbi.nlm.nih.gov/tools/primer-blast/) was used to amplify 200 bp fragment of blaOXA-232 gene. The reaction conditions for the PCR assay were 95˚C for 2 min, 95˚C for 20 sec, 52˚C for 40 sec, 72˚C for 1 min and 72˚C for 5 min. The PCR was carried out for 35 cycles. The volume of a single reaction mixture for the PCR assay was of 25 μl and contained 2X GoTaq® Green Master Mix (12.5 μl, Promega, Madison, USA), primer forward and reverse 1 μl each (10 pmol/μl), DNA template 2 μl (∼100 ng/μl) and nuclease-free water.
Detection of other carbapenemases
The isolates harbouring blaOXA-232 were also investigated for other carbapenemase-encoding genes belonging to class A (blaKPC, blaIMI/NMC and blaSME), class B (blaNDM, blaVIM, blaIMP, blaGIM, blaSIM and blaSPM) and class D (blaOXA-23, blaOXA-24/40, blaOXA-48, blaOXA-58, blaOXA-134, blaOXA-143, blaOXA-198, blaOXA-211, blaOXA-213, blaOXA-214, blaOXA-228 and blaOXA-211) carbapenemases through multiple PCR assays using previously described primer pairs and reaction conditions22.
Detection of co-existing resistance determinants
The carriage of ESBL genes (blaTEM, blaCTX-M, blaSHV, blaOXA-2, blaOXA-10, blaPER, blaGES and blaVEB), aminoglycoside modifying enzyme (AMEs) genes (ant(2″)-Ia, ant(3″)-I, ant(4′)-Ia, aac(3)-I, aac(3)-IIc, aac(6′)-Ib, aac(6′)-II, aph(2″)-Ib, aph(2″)-Ic, aph(2″)-Id, aph(3′)-I, aph(3′)-IIb, aph(3′)-IIIa, aph(3′)-VI a, and aph(4)-Ia), acquired 16S methyltransferase genes (rmtA, rmtB, rmtC, rmtD, armA and npmA), sulphonamides resistance genes (sul1, sul2 and sul3) and quinolones resistance genes (qnrA, qnrB, qnrC, qnrS, qnrD, qep and aac(6’)-lb-cr) were also investigated in the study isolates through multiple PCR assays which were performed using primer pairs and reaction conditions described earlier23.
Horizontal gene transferability assay
As per manufacturer’s instructions, plasmid from blaOXA-232 harbouring isolates was extracted using QIAprep Spin Miniprep Kit (Qiagen, Germany). The transformation assay was carried out by the heat-shock method using E. coli DH5α as a recipient strain24. The blaOXA-232 transformants were selected on Luria Bertani agar (HiMedia Laboratories Pvt. Ltd.) containing 0.5 µg/ml of imipenem (Merck, France). Conjugation experiments were performed to assess the self-transferability of blaOXA-232. E. coli J53 (sodium azide resistant) was used as recipient strain for the experiment. Liquid-mating assay was performed using the donor and recipient cultures and the transconjugants were selected on Luria Bertani agar (HiMedia Laboratories Pvt. Ltd.) medium supplemented with 0.5 µg/ml of imipenem (Merck, France) and 100 µg/ml of sodium azide (HiMedia Laboratories Pvt. Ltd.)25.
Incompatibility typing of plasmid harbouring blaOXA-232
Plasmids from blaOXA-232 transformants were extracted and used as template DNA for PCR-Based Replicon Typing (PBRT). Eighteen plasmid incompatibility types (FIA, FIB, FIC, HI1, HI2, I1-Iγ, L/M, N, P, W, T, A/C, K, B/O, X, Y, F and FIIA) were targeted through multiple PCR assays using previously described primer pairs and reaction conditions26.
Cloning of blaOXA-232
Cloning was performed with the primer pair OXA-232WF (5/-GCAGTTTGCTAGGGAATGAGAGG-3/) and OXA-232WR (5/-GCATCAGCATTTTGTCCA TATAC-3/) designed using the NCBI primer blast tool (https://www.ncbi.nlm.nih.gov/tools/primer-blast/) from the flanking region of the blaOXA-232 gene to amplify the whole gene (957 bp) including its native promoter region. For amplification, 50 μl reaction volume was used containing 25 μl of 2X GoTaq® Green Master Mix (Promega, Madison, USA), 1 μl of each primer (10 pmol/μl), 2 μl of DNA template (∼100 ng/μl) and nuclease-free water. The amplification conditions included a 2 minutes initial denaturation at 95˚C followed by 35 cycles of 95˚C for 20 seconds, 52˚C for 40 seconds and 72˚C for 1 minute with a single final extension cycle of 72˚C for 5 minutes.
The amplified PCR products were then purified using the MinElute® PCR Purification Kit (Qiagen, Hilden, Germany) and ligated into pMD20-T vector as per the manufacturer’s protocol (Mighty TA-cloning Kit, TaKaRa, Japan). The recombinant plasmid was then transformed into E. coli DH5α by the heat-shock method and the clones were selected by blue-white screening on LB agar medium (HiMedia Laboratories Pvt. Ltd.) containing ampicillin (100 μg/mL) and imipenem (0.25 μg/ml) and were further confirmed by colony PCR.
Antibiogram of clones
The susceptibility of the blaOXA-232 clones towards ertapenem, imipenem and meropenem were investigated via Kirby-Bauer Disc Diffusion method and BMD following the CLSI 2022 guidelines20.
Transcriptional expressional of blaOXA-232 under carbapenem stress
For expression study, blaOXA-232 harbouring isolates and their corresponding clones were grown in LB broth (HiMedia Laboratories Pvt. Ltd.) containing 0.5 μg/mL and 1 μg/ml ertapenem, imipenem and meropenem. In accordance with the manufacturer’s instructions, mRNA was extracted using Qiagen RNeasy Mini Kit (Qiagen, Hilden, Germany) followed by preparation of cDNA and its quantification via QuantiTect® Reverse Transcription Kit (Qiagen, Hilden, Germany) and Picodrop (Pico 200, Cambridge, UK), respectively. To analyse the expression level blaOXA-232 gene under carbapenem stress, quantitative Real-Time PCR (qRT-PCR) was performed in triplicate form in StepOnePlus™ Real-Time PCR system (Applied Biosystems, USA) using PowerUp™ SYBR® green PCR Master Mix (Applied Biosystems, USA) and the primer pair OXA-232RTF (5/-CCCAATAGCTTGATCGCCCT-3/) and OXA-232RTR (5/-TATCACGCGTCTGTCCATCC-3/) was designed using the NCBI primer blast tool (https://www.ncbi.nlm.nih.gov/tools/primer-blast/). The relative fold change in the expression level of the blaOXA-232 gene was measured by the ΔΔCT method, and Ct value of each test sample was normalised against the housekeeping gene pmrA used as an endogenous control for the reaction. Relative quantification (RQ) values of blaOXA-232 gene were compared with the reference sample E. coli ATCC 25922 without carbapenem stress and after exposure to varying concentrations of carbapenems.
Association of blaOXA-232 with mobile genetic elements
To investigate the linkage of blaOXA-232 with the insertion sequences ISAba1, ISAba4, ISAba125 and ISEcp1, four sets of PCR assay were performed using the forward primers of ISAba1 (5/-CGACGAATACTATGACAC-3/), ISAba4 (5/-ACTCTCATATTTTTTCTTGG-3/), ISAba125 (5/-GAAACTGTCGCACCTCATGTTTG-3/) and ISEcp1 (5/-TTCAAAAAGCATAATCAAAGCC-3/) and the reverse primer of blaOXA-232 (5/-AGGGCGATCAAGCTATTGGG-3/)27,28. The reaction was performed in T100TM Thermal cycler (Bio-Rad, USA) and a single PCR reaction mixture was of 50 μl volume and contained 2X GoTaq® Green Master Mix (25 μl, Promega, Madison, USA), primer forward and reverse 2 μl each (10 pmol/μl), DNA template 2 μl (∼100 ng/μl) and nuclease-free water. The amplification conditions included an initial denaturation of 2 min at 95˚C followed by 34 cycles of 95˚C for 20 sec, 44˚C for 45 sec and 72˚C for 2 min with a single final extension cycle of 72 ˚C for 7 min. The amplified PCR products were then sequenced, and the results were analysed using NCBI BLAST suite programme (http://blast.ncbi.nlm.nih.gov/Blast.cgi).
Typing of isolates by ERIC PCR
The clonal relatedness of the blaOXA-232 harbouring isolates was determined by ERIC PCR using the universal primers ERIC-F (5/-ATGTAAGCTCCTGGGGATTCAC-3/) and ERIC-R (5/-AAGTAAGTGACTGGGGTGAGCG-3/). The PCR assay was carried out using amplification conditions described previously29. A dendrogram was created using NTSYS-pc version 2.0 software based on the banding patterns of the amplified products observed in agarose gel through Gel Doc™ EZ Gel Documentation System (Bio-Rad, California, USA).
Results
Carbapenem resistance in the isolates
All 35 K. pneumoniae isolates, comprising 14 isolates from blood and 21 isolates from anal swab culture surveillance were phenotypically confirmed for carbapenemase production by Carba NP test. The study isolates exhibited resistance towards all the antibiotics used in antibiotic susceptibility testing (Table I). MICs against carbapenem group of antibiotics were above breakpoint (≥64 µg/ml).
| Isolates ID | Antimicrobial resistance profile | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CPM | CRO | CAZ | ATM | IPM | MEM | ETP | PIT | AMK | GEN | CIP | LVX | |
| BKp1 | R | R | R | R | R | R | R | R | R | R | R | R |
| BKp4 | R | R | R | R | R | R | R | R | R | R | R | R |
| BKp5 | R | R | R | R | R | R | R | R | R | R | R | R |
| BKp8 | R | R | R | R | R | R | R | R | R | R | R | R |
| BKp15 | R | R | R | R | R | R | R | R | R | R | R | R |
| BKp18 | R | R | R | R | R | R | R | R | R | R | R | R |
| BKp22 | R | R | R | R | R | R | R | R | R | R | R | R |
CPM, cefepime (30 µg); CRO, ceftriaxone (30 µg); CAZ, ceftazidime (30 µg); ATM, aztreonam (30 µg); IPM, imipenem (10 µg); MEM, meropenem (10 µg); ETP, ertapenem (10 µg); PIT, piperacillin/tazobactam (100/10 µg); AMK, amikacin (10 µg); GEN, gentamicin (10 µg); CIP, ciprofloxacin (5 µg); LVX, levofloxacin (5µg); R, resistant
Carbapenemase genes in the isolates
PCR assay detected seven K. pneumoniae isolates harbouring blaOXA-232 gene. In these seven isolates, no other carbapenemase genes were detected. All seven isolates were from blood samples of neonates isolated from different outbreaks of CRKP in the NICU.
Co-carriage of other genes
Additionally, co-carriage of different resistance genes was observed in the blaOXA-232 harbouring isolates. In one isolate, a combination of blaCTX-M-15, qnrB and qnrD genes was detected, while in another isolate, co-carriage of blaCTX-M-15, sul1 and sul2 was observed. In another isolate, blaCTX-M-15, along with, sul1 was detected, while blaCTX-M-15 and sul2 were identified in two isolates. One isolate each was co-harbouring blaTEM and aac(6’)lb-cr along with blaOXA-232. The profile of these isolates are summarised in Table II.
| Isolates ID | Carbapenemase | Other co-resistance | ERIC type |
|---|---|---|---|
| BKp1 | blaOXA-232 | blaCTX-M-15, sul2 | I |
| BKp4 | blaOXA-232 | aac(6’)lb-cr | I |
| BKp5 | blaOXA-232 | blaCTX-M-15, sul1, sul2 | I |
| BKp8 | blaOXA-232 | blaTEM | I |
| BKp15 | blaOXA-232 | blaCTX-M-15, qnrB, qnrD | II |
| BKp18 | blaOXA-232 | blaCTX-M-15, sul2 | I |
| BKp22 | blaOXA-232 | blaCTX-M-15, sul2 | I |
ERIC, enterobacterial repetitive intergenic consensus
Horizontal gene transfer analysis and cloning of blaOXA-232
Transformants harbouring plasmid with blaOXA-232 were successfully selected on the medium containing imipenem and were confirmed by PCR assay. However, the gene was located on a non-conjugative plasmid as the conjugation experiment could not produce any transconjugant. Analysis of transformants via PBRT for replicon types yielded no results. Cloning of blaOXA-232 was successful, which was confirmed by colony PCR. The clones were non-susceptible towards ertapenem, imipenem and meropenem.
Expression studies of blaOXA-232 level
Analysis of qRT-PCR results revealed that the blaOXA-232 level was increased by two-folds and three-folds in the isolates under sub-inhibitory concentrations of imipenem and ertapenem, respectively. While in their clones, a substantial escalation in the transcriptional level of blaOXA-232 was observed under sub-inhibitory concentrations of these carbapenems. There was an increase in the levels of blaOXA-232 in the isolates with increase in meropenem sub-inhibitory concentrations. However, in the clones, the expression decreased with an increase in meropenem concentration. The average relative quantification values (2-ΔΔCT) of the isolates harbouring blaOXA-232 and their respective clones against sub-inhibitory concentrations of imipenem, ertapenem, and meropenem have been summarized in Figure 1 and Supplementary Table I.
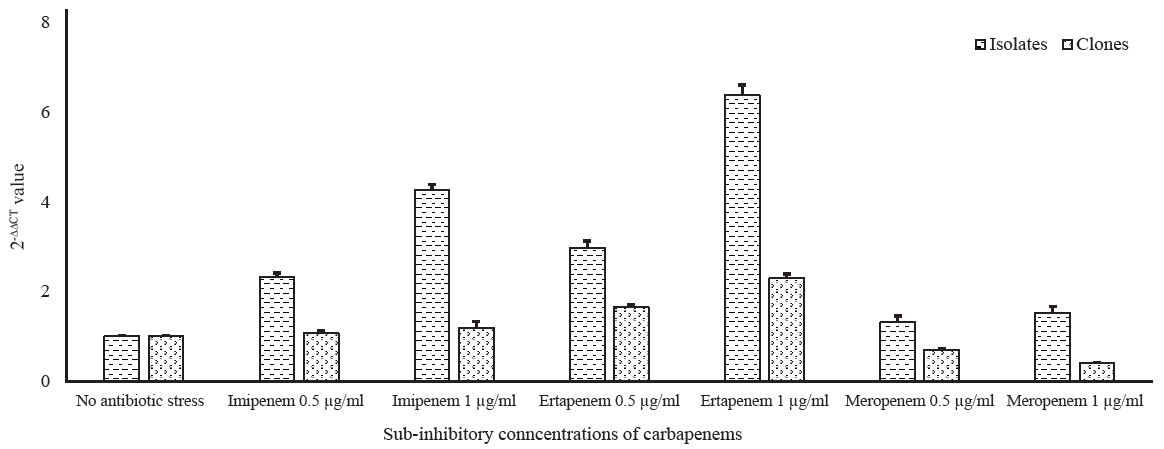
- Transcriptional expression of blaOXA-232 in wild types and clones under sub-inhibitory pressure of carbapenems.
Insertion sequence and DNA fingerprinting
Genetic analysis of blaOXA-232 revealed the association with the insertion sequence ISEcp1. In all seven blaOXA-232 harbouring isolates, the gene was found to be associated with ISEcp1 in the upstream region. Two different haplotypes of K. pneumoniae were identified in DNA fingerprinting by ERIC PCR, which were associated with the carriage and dissemination of the blaOXA-232 gene (Figure 2 and Supplementary Table II).

- Clonal relatedness of Klebsiella pneumoniae isolates harbouring blaOXA-232.DAS et al: CHARACTERIZATION OF BLAOXA-232 CARRYING CRKP
Discussion
This study reported seven CRKP with the carriage of blaOXA-232, which is presently the dominant variant of OXA-48 like carbapenemases in Southeast Asian countries. OXA-48-like carbapenemases are considered endemic to countries in the Middle East, Mediterranean regions and the Indian subcontinent30. The first report of OXA-232 from India was in 2019. The gene was identified in diverse sequence types of K. pneumoniae isolates of clinical origin and was found localized on ColKP3- and ColE-type plasmids12. In 2020, a hypervirulent K. pneumoniae belonging to ST23 associated with neonatal sepsis was identified with the carriage of blaOXA-23217. In line with the previous studies, we also noticed K. pneumoniae harbouring blaOXA-232 exhibiting a high level of resistance towards carbapenems and other antibiotics. Several instances of K. pneumoniae harbouring OXA-232 and co-producing blaNDM-1 have been reported previously10,14,31. However, in the present investigation, no other carbapenemase genes in blaOXA-232 harbouring isolates were found, which were in agreement with studies conducted earlier12,13,15-18. The most probable reason for the absence of other carbapenemase genes could be due to the reduction in fitness cost of these isolates as previous studies already found that isolates co-harbouring blaOXA-232 with metallo-β-lactamase blaNDM had different plasmids carrying the carbapenemase gene14. As these isolates were also co-carrying other multiple resistant genes, it could affect their fitness. Additionally, in these isolates, OXA-232 expression was associated with high-level carbapenem resistance; therefore, added carbapenem mechanisms were not required. Further studies should confirm these speculations, and were not within the scope of the present investigation. Otherwise, there has been a recent shift in the blaOXA-48 endemicity from the existing blaNDM, thus accounting for their predominance12.
K. pneumoniae harbouring blaOXA-232 with co-carriage of ESBLs, aminoglycoside-modifying enzymes, sulphonamides and quinolone resistance genes were described in several studies conducted earlier. Mukherjee et al17 in their study reported occurrence of blaOXA-232 in hypervirulent K. pneumoniae co-producing blaTEM, blaCTX-M-15, rmtF, armA, aph(3”)-Ib, sul1, sul2 and aac(6’)-Ib-cr. Similarly, Weng and his team 18 in 2020 reported the co-carriage of blaTEM, blaCTX-M, aph(3)-IIa, aph(3”)-Ib, aph(6)-Id and rmtD in blaOXA-232 harbouring K. pneumoniae18. In our study, we also observed the occurrence of blaCTX-M-15, blaTEM, blaCTX-M-15, sul1, sul2, qnrB, qnrD and aac(6’)-Ib-cr among the blaOXA-232 positive isolates. The presence of such diverse genes conferring resistance to a wide range of antibiotics warrants a major concern to public health as it significantly limits therapeutic options. In our study, the transformation of plasmid with blaOXA-232 was successfully carried out, though the conjugation assay was unsuccessful. This might be due to the carriage of this resistance determinant within the non-conjugative plasmid in the studied isolates. This assumption is supported by various previous findings where the gene was located in a non-conjugative plasmid11,16-18.
In this study, sub-inhibitory concentrations of carbapenems were used to determine the effects of selective carbapenem pressure on the expression of blaOXA-232 in the isolates and their clones. It was revealed that sub-inhibitory concentrations of ertapenem, imipenem and meropenem had significant effects on the expressional level of blaOXA-232 when compared with the unexposed isolates as well as their respective clones. This finding implies that selective carbapenem pressure induces an adaptive response and contributes to the elevated expression of this resistance determinant thereby adding to carbapenem resistance. As heavy carbapenem use in hospitals, especially in developing countries, is already an established risk factor for the emergence of carbapenem-resistant organisms, the selective pressure thus caused could result in such adaptations in these isolates32. Studies have revealed the association of diverse insertion sequences and transposons with blaOXA-23212. In our study, the insertion sequence ISEcp1 was found associated with blaOXA-232, emphasising the role of this mobile genetic element in the maintenance and dissemination of this carbapenem resistance determinant within the two haplotypes of K. pneumoniae. Though clonal expansion with the determination of sequence types could not be done in this study, it was revealed that transmission of these isolates could be facilitated by this insertion sequence under antibiotic pressure in the hospital environment.
Overall, the study characterised blaOXA-232-carrying CRKP isolates of clinical origin and clearly demonstrated that higher expression of this gene occurred under sub-inhibitory antibiotic pressure, which could lead to inducible resistance during therapy. With the emergence of further reports on this carbapenemase variant and its adaptations against various factors in the hospital environment, urgent monitoring of this variant should be done to understand its transmission dynamics and to limit further dissemination.
Financial support & sponsorship
This research work was carried out with the support of the funding agencies; Department of Biotechnology (DBT), Government of India (BT/PR242/NER/98/716/2017) and Indian Council of Medical Research, India (ICMR), through Senior Research Fellowship (ICMR-SRF) (2020-7955/CMB-BMS) awarded to the first author (BJD).
Conflicts of Interest
None.
Use of Artificial Intelligence (AI)-Assisted Technology for manuscript preparation
The authors confirm that there was no use of Artificial Intelligence (AI)-Assisted Technology for assisting in the writing of the manuscript and no images were manipulated using AI.
References
- An outbreak of carbapenem-resistant and hypervirulent Klebsiella pneumoniae in an intensive care unit of a major teaching hospital in Wenzhou, China. Front Public Health. 2019;7:229-40.
- [Google Scholar]
- Bloodstream infections caused by carbapenemase-producing Klebsiella pneumoniae among intensive care unit patients after orthotopic liver transplantation: risk factors for infection and impact of resistance on outcomes. Transplant Proc. 2014;46:3216-18.
- [Google Scholar]
- Occult Klebsiella pneumoniae bacteremia at emergency department: A single center experience. J Microbiol Immunol Infect. 2015;48:684-91.
- [Google Scholar]
- Risk factors for carbapenem-resistant Klebsiella pneumoniae infection and mortality of Klebsiella pneumoniae infection. Chin Med J (Engl). 2018;131:56-62.
- [Google Scholar]
- WHO publishes list of bacteria for which new antibiotics are urgently needed. Available from: https://www.who.int/news/item/27-02-2017-who-publishes-list-of-bacteria-for-which-new-antibiotics-are-urgently-needed, accessed on July 11, 2022.
- Indian Priority pathogen list: to guide research, discovery, and development of new antibiotics in India. Available from: https://dbtindia.gov.in/sites/default/files/IPPL_final.pdf, accessed on July 10, 2022.
- Extensively drug-resistant hypervirulent Klebsiella pneumoniae from a series of neonatal sepsis in a tertiary care hospital, India. Front Med (Lousanne). 2021;8:645955-7.
- [Google Scholar]
- Extensive outbreak of colistin resistant, carbapenemase (blaOXA-48, blaNDM) producing Klebsiella pneumoniae in a large tertiary care hospital, India. Antimicrob Resist Infect Control. 2022;11:1-9.
- [Google Scholar]
- Occurrence of blaNDM variants among enterobacteriaceae from a neonatal intensive care unit in a Northern India hospital. Front Microbiol. 2018;9:407-17.
- [Google Scholar]
- Co-production of NDM-1 and OXA-232 by ST16 Klebsiella pneumoniae, Italy, 2016. Future Microbiol. 2017;12:1119-22.
- [Google Scholar]
- Genetic and biochemical characterisation of OXA-232, a carbapenem-hydrolysing class D β-lactamase from Enterobacteriaceae. Int J Antimicrob Agents. 2013;41:325-9.
- [Google Scholar]
- Rapidly disseminating blaOXA-232 carrying Klebsiella pneumoniae belonging to ST231 in India: multiple and varied mobile genetic elements. BMC Microbiol. 2019;19:137-45.
- [Google Scholar]
- Emergence of clinical Klebsiella pneumoniae producing OXA-232 carbapenemase in Singapore. New Microbes New Infect. 2013;1:13-5.
- [Google Scholar]
- Co-production of NDM-1 and OXA-232 by Klebsiella pneumoniae. Emerg Infect Dis. 2014;20:163-5.
- [Google Scholar]
- Clonal dissemination of OXA-232 carbapenemase-producing Klebsiella pneumoniae in neonates. Antimicrob Agents Chemother. 2017;61:e00385-17.
- [Google Scholar]
- Prevalence and phenotypic characterization of carbapenem-resistant Klebsiella pneumoniae strains recovered from sputum and fecal samples of ICU patients in Zhejiang Province, China. Infect Drug Resist. 2018;12:11-18.
- [Google Scholar]
- Emergence of OXA-232-producing hypervirulent Klebsiella pneumoniae ST23 causing neonatal sepsis. J Antimicrob Chemother. 2020;75:2004-2006.
- [Google Scholar]
- The Characterization of OXA-232 Carbapenemase-Producing ST437 Klebsiella pneumoniae in China. Can J Infect Dis Med Microbiol 2020:20205626503-5.
- [Google Scholar]
- Increased plasmid copy number contributes to the elevated carbapenem resistance in OXA-232-producing Klebsiella pneumoniae. Microb Drug Resist. 2020;26:561-68.
- [Google Scholar]
- Performance Standards for Antimicrobial Susceptibility Testing. Available from: https://clsi.org/media/wi0pmpke/m100ed32_sample.pdf, accessed on July 11, 2022.
- Evaluation of different DNA extraction procedures for the detection of Salmonella from chicken products by polymerase chain reaction. Lett Appl Microbiol. 1994;19:294-8.
- [Google Scholar]
- Propagation of blaKPC-2 within two sequence types of Escherichia coli in a tertiary referral hospital of northeast India. Gene Rep. 2021;24:101283-10.
- [Google Scholar]
- Occurrence of blaOXA-48 type carbapenemase in Escherichia coli with coexisting resistance determinants: A report from India. Gene Rep. 2022;26:101459.
- [Google Scholar]
- Elimination of diverse Inc type plasmids carrying carbapenemase genes within Escherichia coli of clinical origin: A single-center study from North-east India. Gene Rep. 2023;31:101770-9.
- [Google Scholar]
- Identification of plasmids by PCR-based replicon typing. J Microbiol Methods. 2005;63:219-28.
- [Google Scholar]
- Genetics and expression of the carbapenem-hydrolyzing oxacillinase gene blaOXA-23 in Acinetobacter baumannii. Antimicrob Agents Chemother. 2007;51:1530-33.
- [Google Scholar]
- blaNDM-1 is a chimera likely constructed in Acinetobacter baumannii. Antimicrob Agents Chemother. 2012;56:2773-76.
- [Google Scholar]
- Distribution of repetitive DNA sequences in eubacteria and application to fingerprinting of bacterial genomes. Nucleic Acids Res. 1991;19:6823-31.
- [Google Scholar]
- Global dissemination of carbapenemase-producing Klebsiella pneumoniae: Epidemiology, genetic context, treatment options, and detection methods. Front Microbiol. 2016;7:895-925.
- [Google Scholar]
- Coinfections of two strains of NDM-1- and OXA-232-coproducing Klebsiella pneumoniae in a kidney transplant patient. Antimicrob Agents Chemother. 2020;64:e00948-19.
- [Google Scholar]
- A Systematic review and meta-analyses of the clinical epidemiology of carbapenem-resistant enterobacteriaceae. Antimicrob Agents Chemother. 2017;62:e01730-17.
- [Google Scholar]






