Translate this page into:
Characterization of antimicrobial resistance markers & their stability in Salmonella enterica serovar Typhi
Reprint requests: Dr. Yashwant Kumar, National Salmonella & Escherichia Centre, Central Research Institute, Kasauli 173 204, Himachal Pradesh, India e-mail: yasht26@yahoo.co.in
-
Received: ,
This is an open access article distributed under the terms of the Creative Commons Attribution-NonCommercial-ShareAlike 3.0 License, which allows others to remix, tweak, and build upon the work non-commercially, as long as the author is credited and the new creations are licensed under the identical terms.
This article was originally published by Medknow Publications & Media Pvt Ltd and was migrated to Scientific Scholar after the change of Publisher.
Abstract
Background & objectives:
Typhoid fever is a major cause of morbidity and mortality in the developing countries including India. Resistance to multiple antimicrobial agents is an emerging global problem that has serious impact on the treatment of disease. There are many factors associated with the emergence of resistance. Most important of them is the acquisition and further transmission and spread of resistance markers among various bacterial species. Therefore, we conducted this study to characterize the resistance plasmids in terms of their transferability and stability among Salmonella enterica serovar Typhi.
Methods:
Six multidrug-resistant S. Typhi isolates were evaluated for the stability and transfer of resistance markers. The resistance plasmids were also checked for the presence of RepHI1A replicon.
Results:
All resistance markers were found to be transferred to the recipient through conjugation and transformation, except for nalidixic acid. None of the resistance plasmid was found to harbour RepHI1A replicon and therefore, did not belong to incompatibility group IncHI1. Resistance markers were found to be highly stable in all the isolates during serial passages and storage as stab cultures at different temperatures for different time periods.
Interpretation & conclusions:
Resistance markers for chloramphenicol, ampicillin, streptomycin and trimethoprim were transferred through conjugation as well as transformation whereas that for nalidixic acid was not transferred in any of the isolates. Markers for chloramphenicol and streptomycin resistance were found to be most stable during various storage conditions. Presence of small-sized non-IncHI1 resistance plasmids is a matter of concern due to their capability to exist inside the host, thereby increasing the possibility of their transmission and spread among S. Typhi and other bacterial species.
Keywords
Resistance plasmids
Salmonella Typhi – stability
transmission
Typhoid fever remains a major public health threat globally with over 21.6 million cases and 220,000 deaths occurring annually12. Although typhoid fever is not common in the developed countries due to improved sanitation and hygiene, it remains an important and persistent health problem in the developing nations with highest incidence in India and Pakistan34.
Emergence of antimicrobial resistance among Salmonella enterica serovar Typhi (S. Typhi) is a major problem. Being the most effective drug, chloramphenicol was used for the treatment of typhoid fever until the emergence of chloramphenicol-resistant strains of S. Typhi in 197256. Since 1989, emergence of multidrug-resistant (MDR) S. Typhi, resistant to chloramphenicol, ampicillin, trimethoprim, streptomycin, sulphamethoxazole and tetracycline, has been reported from the developing countries including India7-9. Therefore, fluoroquinolone class of antibiotics became the treatment of choice for enteric fever1011.
S. Typhi strains those were resistant to nalidixic acid and exhibited reduced susceptibility to quinolones have been reported subsequently in a number of countries12. There have been several reports indicating the re-emergence of susceptibility to conventionally used drugs1314, which raise the possibility of their reintroduction into therapeutic regimen. Among S. Typhi, MDR is associated with self-transmissible IncHI1 plasmids, which carry a panel of genes conferring resistance to several first-line antimicrobials15. From 1995 onwards, 98 per cent of MDR S. Typhi isolates were of haplotype H58, carrying the IncHI1 plasmid16 and are widespread in Africa and Asia17. In view of these observations, this study was undertaken to characterize the antimicrobial resistance in S. Typhi isolates in terms of the resistance markers which render the microorganism resistant to different classes of antibiotics.
Material & Methods
In our previous study18, of the 128 isolates of S. Typhi received during the 2008-2009 at the National Salmonella and Escherichia Centre, Central Research Institute, Kasauli, India, from all around the country, six were found to be MDR showing resistance to chloramphenicol, ampicillin, streptomycin, nalidixic acid and trimethoprim (C-A-S-Na-Tr), the MDR isolates were used in the present study for characterization of resistance markers. These were tested for the transfer of resistance through conjugation and transformation. The resistance plasmids were further tested to determine if they belonged to incompatibility group IncHI. Stability of resistance markers was also assessed during regular subcultures and storage at different temperatures for different time period.
Conjugation and transformation: Conjugation experiments were performed according to the method previously reported19 to check the transfer of resistance from donor (each S. Typhi isolate) to recipient [CGSC 6576 (F−, λ−, recA1), IN(rrnD-rrnE)1, rpoB331 (rifampicin resistance [RifR]), hsdR19], (provided by Dr John Wertz, Coli Genetic Stock Centre, Yale University, New Haven, CT, USA). For transformation, plasmid DNA was isolated from each S. Typhi isolate as previously reported20. Transformation experiments were carried out using Escherichia coli HMS174 (F- recA1 hsdR (rK12− mK12+) (RifR) competent cell kit (Novagen, Germany) as per the manufacturer’s instructions.
PCR for RepHI1A replicon: Plasmid DNA isolated from each isolate was tested for the presence of RepHI1A replicon by PCR using the primers 5’-GGTCCAACCCATTGCTTTAC-3’ and 5’-CACG GAAAGAAATCACAAC-3’ as previously reported21 on Mastercycler (Eppendorf AG, Germany). Briefly, PCR reaction was set up using 50 ng plasmid DNA and 50 nM of each primer in a buffer composed of 10 mM Tris-HCl (pH 8.3), 50 mM KCl, 1.5 mM MgCl2, 200 µM dNTP mixture and 1U of Taq polymerase in a final volume of 100 µl. Amplification conditions were thirty cycles of 94°C/30 sec, 55°C/30 sec and 72°C/30 sec, with a final extension step of 72°C for 10 min. The gel was observed for 365 bp amplicon suggestive of RepHI1A replicon as reported previously22.
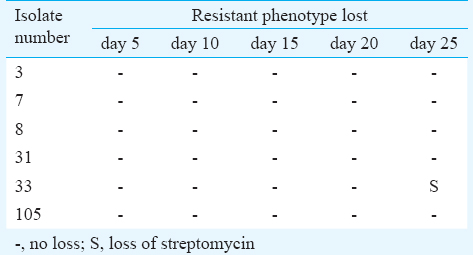
Stability of resistance plasmids during subcultures: Stability of the resistance markers in the test strains was determined as described23. The resistance pattern and the plasmid content were determined before conducting the study. Briefly, each isolate was sub-cultured in 5 ml of pen assay broth (Becton-Dickinson, New Jersey, USA) and incubated at 37°C for overnight. Daily sub-culture was performed in fresh pen assay broth for 25 days, and stability of resistance pattern was determined intermittently on 5, 10, 15, 20 and 25 passage by antimicrobial susceptibility test by disc diffusion method24.
Stability of resistance markers during storage: Stability of the resistance markers in each isolate was determined as described previously25 after storage at 4°C, 22°C and 30°C for 1, 3, 6, 9 and 12 months. Each isolate was inoculated in Luria-Bertani (LB) broth and incubated at 37°C. From this, a series of identical stab cultures were prepared in Difco agar. The stab cultures were sealed with a cork soaked in heated, sterile Vaseline. These were stored at 4, 22 and 30°C. At varying intervals (after 1, 3, 6, 9 and 12 months), stab cultures were revived by inoculating in 0.5 ml of fresh LB broth and incubated overnight at 37°C and tested for any loss of resistance marker by disc diffusion susceptibility testing.
Results
All six isolates of S. Typhi transferred resistance markers for chloramphenicol, ampicillin, streptomycin and trimethoprim to the recipient by conjugation as well as transformation. However, resistance marker for nalidixic acid was not transferred from any of the tested isolates by either of the methods (Table I).

All isolates were found to harbour a single plasmid of about 33.5 kb. During the detection of RepHI1A replicon, none of the isolates exhibited the presence of the same. Positive control (plasmid from strain CT18) produced a fine band of 365 bp showing the presence of RepHI1A replicon (Figure).
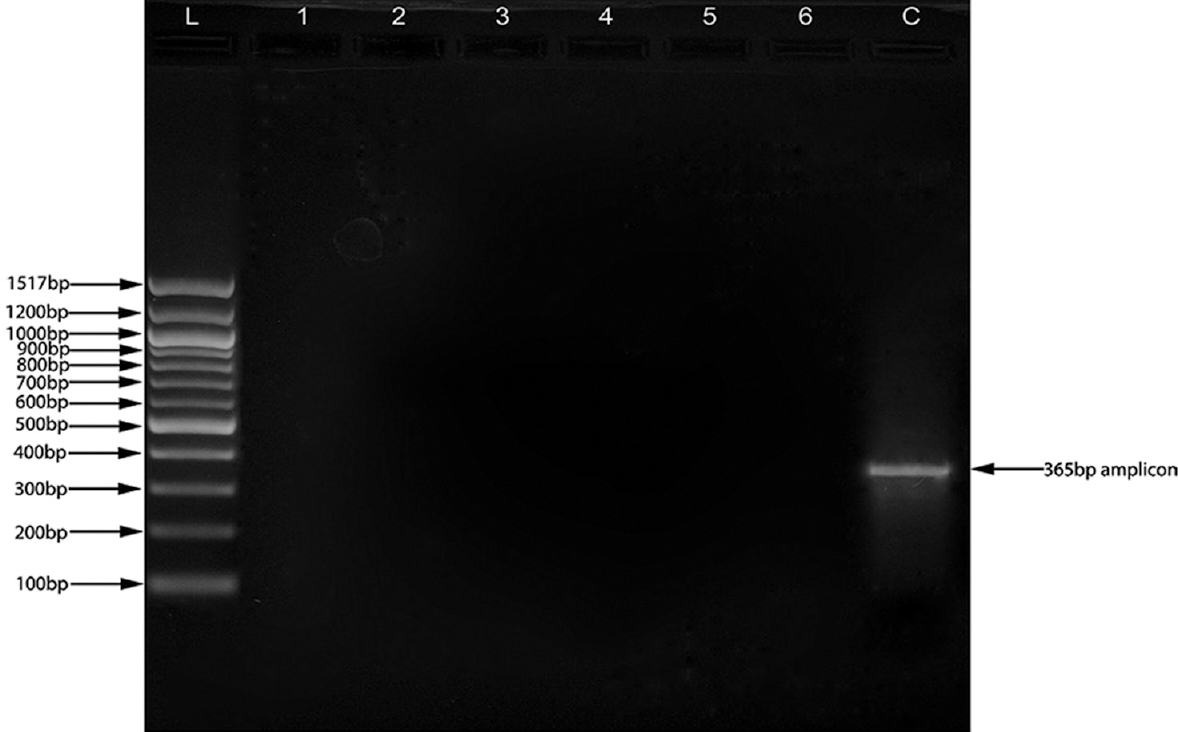
- Polymerase chain reaction for the detection of RepH1A replicon. All six isolates were found to be negative for the RepH1A replicon, whereas plasmid from the Salmonella enterica serovar Typhi CT18 produced a fine band of 365 bp representative of RepH1A replicon. L: Ladder, Lane 1-6: Test plasmid DNA, C: plasmid from Salmonella enterica serovar Typhi CT18.
No loss of resistance markers was observed in 83.3 per cent of the isolates during serial passages up to 25th passage, except for loss of streptomycin resistance in one isolate at 25th passage (Table II). During storage of the isolates at different temperatures for different time periods, streptomycin was lost in one isolate on 12 months storage at 2-8°C whereas chloramphenicol and streptomycin were lost on 12 months storage at 30°C in another (Table III).

Discussion
MDR in S. enterica serovar Typhi poses a major threat to the effective treatment of the disease and has been prevalent in India and the neighbouring countries since 19892627. In most cases, MDR is associated with high molecular weight and self-transferable plasmid transferring the phenotype individually or en bloc2829.
In the present study, of five resistance markers, only four were found to be transferred both through conjugation and transformation. Nalidixic acid resistance marker was not transferred by either of the methods. Transfer of resistance phenotype through conjugation and transformation indicated plasmid-borne antibiotic resistance. The absence of transfer of nalidixic acid resistance may be due to its chromosomal origin30. Quinolone resistance is mediated by non-transmissible, spontaneously occurring point mutations in chromosomal genes (gyrA, gyrB, parC and parE). These point mutations alter the enzymes, DNA gyrase and topoisomerase IV that are targets for quinolone drugs. The most frequent mutation is a single-point mutations in the gyrA gene, characteristically occurring at position 83 of the DNA gyrase enzyme (changing serine to phenylalanine) and position 87 (changing aspartate to tyrosine or glycine)3132.
The mechanism of fluoroquinolone resistance in S. enterica serovar Typhi is not completely understood and there have been only a few studies on fluoroquinolone resistance in this organism33. A single mutation in the gyrA gene is sufficient to confer resistance to nalidixic acid and reduced susceptibility to fluoroquinolones, and a second mutation leads to high level fluoroquinolone resistance3435.
Plasmid-mediated quinolone resistance appears often to be co-transmitted with resistance to broad-spectrum β-lactmases, enabling the promulgation of simultaneous resistance to two classes of antimicrobial agents commonly used to treat serious Salmonella infections36. The present study showed transferrable nature of the MDR, small-sized plasmid through conjugation and transformation, which might contribute to the spread of MDR among S. enterica serovar Typhi, leading to further development of complications in the treatment of typhoid fever.
Plasmids of different incompatibility groups have been found to persist among S. Typhi2937, but those of the incompatibility group IncHI1 appear to be particularly common in this serovar of S. enterica due to its transmission potential38. Plasmids belonging to the incompatibility group IncH1 are frequently the source of resistance to ampicillin, chloramphenicol, trimethoprim and tetracycline. These plasmids are quite large ranging from 140-180 Kb22. Conjugational transfer of these plasmids has been reported to be thermosensitive in nature being transferred at higher frequencies at ambient temperature (27°C) and not at an in vivo temperature (37°C)39.
In the present study, the absence of RepHI1A replicon was shown by non-amplification of the 365 bp region indicative of RepHI1A replicon. This exhibits that the R-plasmids do not belong to the incompatibility group IncH1. This finding is further supported by small size (33.5 kb) of the plasmid and no thermal sensitivity of the plasmid for conjugational transfer. Moreover, S. enterica serovar Typhi strain CT18, containing IncH1 plasmid, was used as a positive control and it gave a band of 365 bp amplicon. Some authors2937 have reported R-plasmids of S. enterica serovar Typhi belonging to the incompatibility group IncA, IncC and IncI. This finding reveals the emergence of non-IncHI1 conjugative plasmids having potential to transfer and spread of MDR among S. enterica serovar Typhi.
Plasmid profiling is a commonly used method of typing bacteria in epidemiological investigations. Therefore, it is pertinent to assess the influence of storage conditions on plasmid stability. Moreover, high stability of R-plasmids ensures the long-term persistence of the same in the bacterium and also increases the possibility of its spread through horizontal transfer.
Resistance plasmids were found to be stable during several serial passages, except loss of streptomycin resistance at 25th passage in one isolate. The same finding of partial loss of resistance due to loss of some of the resistance markers has also been reported previously23. Streptomycin resistance was found to be more prone for loss whereas resistance genes for chloramphenicol, ampicillin and trimethoprim were found to be stable. The presence of highly stable R-plasmids in S. enterica serovar Typhi has also been reported previously from India40. The stable nature of the R-plasmids may increase the chances of conjugational transfer thereby aiding in the spread and maintenance of MDR in S. enterica serovar Typhi. Moreover, due to stable nature of these R-plasmids, plasmid profiling can be used as a reproducible method for epidemiological investigations.
Presence of small-sized, highly stable, non-IncHI1 resistance plasmids among S. enterica serovar Typhi is of major concern due to its transmission potential further increasing the possibility of spread of antimicrobial resistance among this serovar. However, a well-planned and collaborated effort should be made by hospitals, surveillance centres and research institution to keep an eye on the current trends of antimicrobial resistance and the underlying mechanisms.
Acknowledgment
Authors thank the heads of all the laboratories that referred Salmonella isolates to this centre, and Shri Jiwa Ram for supplying media and biochemicals for biotyping. The technical assistance of Sarvshri Gian Chand and Khushnihal Kaushal is also acknowledged.
Conflicts of Interest: None.
References
- Epidemiology of typhoid and paratyphoid fever in India. J Infect Dev Ctries. 2008;2:454-60.
- [Google Scholar]
- A study of typhoid fever in five Asian countries: Disease burden and implications for controls. Bull World Health Organ. 2008;86:260-8.
- [Google Scholar]
- The problem and implications of chloramphenicol resistance in the typhoid bacillus. J Hyg (Lond). 1975;74:289-99.
- [Google Scholar]
- Occurrence and treatment of multiresistant Salmonella typhi. Public Health Lab Serv Microbiol Dig. 1991;8:56-9.
- [Google Scholar]
- Background document: The diagnosis, treatment and prevention of typhoid fever. Geneva, Switzerland: Department of Vaccines and Biologicals, WHO; 2003.
- Changing trends in antimicrobial resistance of Salmonella enterica serovar typhi and Salmonella enterica serovar paratyphi A in Chennai. Indian J Pathol Microbiol. 2009;52:505-8.
- [Google Scholar]
- Antimicrobial susceptibility of Salmonella typhi in India. J Infect Dev Ctries. 2010;4:70-3.
- [Google Scholar]
- Molecular analysis of incHI1 antimicrobial resistance plasmids from Salmonella serovar typhi strains associated with typhoid fever. Antimicrob Agents Chemother. 2003;47:2732-9.
- [Google Scholar]
- Emergence of a globally dominant IncHI1 plasmid type associated with multiple drug resistant typhoid. PLoS Negl Trop Dis. 2011;5:e1245.
- [Google Scholar]
- High-throughput bacterial SNP typing identifies distinct clusters of Salmonella typhi causing typhoid in Nepalese children. BMC Infect Dis. 2010;10:144.
- [Google Scholar]
- Antibiogram profile of Salmonella enterica serovar typhi in India –A two year study. Trop Life Sci Res. 2013;24:45-54.
- [Google Scholar]
- Molecular characterization of antibiotic resistance in clinical Salmonella typhi isolated in Ghana. FEMS Microbiol Lett. 2002;215:249-53.
- [Google Scholar]
- Minipreps of plasmid DNA. In: Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, eds. Current protocols in molecular biology. New York: John Wiley and Sons Inc; 2003. p. :1.6.1-1.6.10.
- [Google Scholar]
- Isolation and location on the R27 map of two replicons and an incompatibility determinant specific for IncHI1 plasmids. J Bacteriol. 1993;175:7697-701.
- [Google Scholar]
- Molecular analysis of and identification of antibiotic resistance genes in clinical isolates of Salmonella typhi from India. J Clin Microbiol. 1998;36:1595-600.
- [Google Scholar]
- Genetic stability of various resistance factors in Escherichia coli and Salmonella typhimurium. J Bacteriol. 1970;102:363-8.
- [Google Scholar]
- Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria That Grow Aerobically; Approved standard-Seventh edition M7-A7. Wayne, PA: CLSI; 2006.
- Stability of plasmids in five strains of Salmonella maintained in stab culture at different temperatures. J Appl Bacteriol. 1994;77:155-9.
- [Google Scholar]
- Multidrug-resistant Salmonella typhi in Haryana in 1989-90. Indian J Med Res. 1992;95:12-3.
- [Google Scholar]
- Multidrug resistant Salmonella typhi in Calicut, South India. Indian J Med Res. 1992;95:68-70.
- [Google Scholar]
- Complete genome sequence of a multiple drug resistant Salmonella enterica serovar typhi CT18. Nature. 2001;413:848-52.
- [Google Scholar]
- Analysis of plasmid and chromosomal DNA of multidrug-resistant Salmonella enterica serovar typhi from Asia. J Clin Microbiol. 2000;38:1449-52.
- [Google Scholar]
- Molecular characterization of ciprofloxacin-resistant Salmonella enterica serovar typhi and paratyphi A causing enteric fever in India. J Antimicrob Chemother. 2006;58:1139-44.
- [Google Scholar]
- Antimicrobial resistance in Salmonella enterica serovar typhi isolates from Bangladesh, Indonesia, Taiwan, and Vietnam. Antimicrob Agents Chemother. 2014;58:6501-7.
- [Google Scholar]
- Mechanisms of antibiotic resistance in Salmonella typhi. Int J Curr Microbiol Appl Sci. 2014;3:461-76.
- [Google Scholar]
- Reduced susceptibility to ciprofloxacin and gyrA gene mutation in North Indian strains of Salmonella enterica serotype typhi and serotype paratyphi A. Microb Drug Resist. 2004;10:146-53.
- [Google Scholar]
- Emergence of fluoroquinolone resistance in Salmonella enterica serovar typhi in Andaman and Nicobar Islands, India. Indian J Med Res. 2012;136:98-101.
- [Google Scholar]
- Antimicrobial resistance, virulence profiles and molecular subtypes of Salmonella enterica serovars typhi and paratyphi A blood isolates from Kolkata, India during 2009-2013. PLoS One. 2014;9:e101347.
- [Google Scholar]
- Plasmid-mediated quinolone resistance in non-typhi serotypes of Salmonella enterica. Clin Infect Dis. 2006;43:297-304.
- [Google Scholar]
- IncHI plasmids, a dynamic link between resistance and pathogenicity. J Infect Dev Ctries. 2008;2:272-8.
- [Google Scholar]
- Molecular fingerprinting of multidrug-resistant Salmonella enterica serotype typhi. Emerg Infect Dis. 1998;4:317-20.
- [Google Scholar]
- Thermosensitive nature of IncHI1 plasmid transfer. Antimicrob Agents Chemother. 2009;53:2703.
- [Google Scholar]
- Genetic stability of IncH1 and other incompatibility group R-plasmids in Salmonella typhi. Indian J Med Res. 1982;76:512-8.
- [Google Scholar]






