Translate this page into:
Bioinformatics characterization of envelope glycoprotein from Kyasanur Forest disease virus
For correspondence: Dr. Devendra T. Mourya, ICMR-National Institute of Virology, Sus Road, Pashan, Pune 411 021, Maharashtra, India e-mail: dtmourya@gmail.com
-
Received: ,
This is an open access journal, and articles are distributed under the terms of the Creative Commons Attribution-NonCommercial-ShareAlike 4.0 License, which allows others to remix, tweak, and build upon the work non-commercially, as long as appropriate credit is given and the new creations are licensed under the identical terms.
This article was originally published by Medknow Publications & Media Pvt Ltd and was migrated to Scientific Scholar after the change of Publisher.
Abstract
Background & objectives:
Kyasanur Forest disease (KFD) is a febrile illness characterized by haemorrhages and caused by KFD virus (KFDV), which belongs to the Flaviviridae family. It is reported to be an endemic disease in Shimoga district of Karnataka State, India, especially in forested and adjoining areas. Several outbreaks have been reported in newer areas, which raised queries regarding the changing nature of structural proteins if any. The objective of the study was to investigate amino acid composition and antigenic variability if any, among the envelope glycoprotein (E-proteins) from old and new strains of KFDV.
Methods:
Bioinformatic tools and techniques were used to predict B-cell epitopes and three-dimensional structures and to compare envelope glycoprotein (E-proteins) between the old strains of KFDV and those from emerging outbreaks till 2015.
Results:
The strain from recent outbreak in Thirthahalli, Karnataka State (2014), was similar to the older strain of KFDV (99.2%). Although mutations existed in strains from 2015 in Kerala KFD sequences, these did not alter the epitopes.
Interpretation & conclusions:
The study revealed that though mutations existed, there were no drastic changes in the structure or antigenicity of the E-proteins from recent outbreaks. Hence, no correlation could be established between the mutations and detection in new geographical areas. It seems that KFDV must be present earlier also in many States and due to availability of testing system and alertness coming into notice now.
Keywords
B-cell epitopes
ELISA
envelope glycoprotein
Kyasanur Forest disease
phylogenetic analyses
virus
Kyasanur Forest disease (KFD) was first documented as an outbreak in people living in Kyasanur Forest in Karnataka, India12. It is a febrile illness characterized by haemorrhages and is reported to be endemic in Shimoga district of Karnataka2. It is caused by KFD virus (KFDV), which belongs to the Flaviviridae family, transmitted to humans and monkeys by Haemaphysalis ticks. The virus was earlier thought to be related to the Russian spring-summer encephalitis complex of tick-borne viruses1 and shared many characteristics with other flaviviruses. Later on, it was found that though virus was from Flavi genus, it caused viral haemorrhagic fever not encephalitis. The virion is 45 nm in diameter and contains approximately 11 kb genome. The single open reading frame encodes for structural proteins, namely core (C), envelope (E) and membrane protein (M), and seven non-structural proteins, viz. NS1, NS2A, NS2B, NS3, NS4A, NS4B and NS53.
The major envelope glycoprotein E plays an important role in the biology of KFDV. Like other Flavivirus E-proteins, it helps in receptor binding and entry into host cells by the fusion of viral and cellular membrane. However, it also generates host immune responses by inducing protective and neutralizing antibodies. Hence, it is a very important antigenic protein for the development of vaccines and ELISA-based diagnostic assays4. The detection of the disease to newer areas in India567 is a matter of concern and necessitates research to focus on understanding the pathogenesis, development of vaccines and efficient diagnostic kits. Detailed knowledge of KFD pathogenicity, transmission routes and different hosts will help in development of cheap and highly effective diagnostics and enhanced surveillance which will in turn help in reducing disease fatality8910. In addition, increased awareness of forest dwellers as well as travellers is also required to contain this disease.
It is necessary to study the characteristics of the envelope glycoprotein for better understanding of the pathogenesis of KFDV. The E-protein of flaviviruses is a beta-class protein and consists of three domains. The central domain I (DI) connects the extended domain II (DII) with the globular domain III and helps in receptor binding, as known in the case of other flaviviruses such as dengue and Japanese encephalitis, where the B-cell epitopes occur in the domains DI and DII11.
It has been suggested that KFDV shows long range distribution possibly due to widespread movement of birds and the strains of KFDV share a common ancestry12. In the present study, bioinformatic techniques were used to predict B-cell epitopes and to compare E proteins between the old strains and those from emerging outbreaks. In the absence of experimentally known three-dimensional (3D) structure of the protein, in silico homology modelling tools were employed to generate models.
Material & Methods
The study was conducted by considering representative sequences of strains from 1965 to 2014. KFDV E-protein sequences were obtained from the NCBI database (https://www.ncbi.nlm.nih.gov/) and sequences from the recent KFD outbreaks were also included in the analysis. Multiple sequence alignment was performed considering E-protein sequences from tick-borne encephalitis and Japanese encephalitis viruses (TBEV and JEV) as outliers. Pairwise sequence comparisons of the ectodomains for all the possible pairs in the data set were performed1314. Epitope predictions were carried out for selected strains using in silico techniques15. The 3D structure prediction and structural comparison between E-proteins of different KFDV strains were carried out to study if there were any mutations and whether they were affecting the structural proteins.
RNA was extracted from virus isolates using the QIAamp Viral RNA Mini Kit (Qiagen, USA). KFDV E complete gene was amplified using the gene-specific primers. Primer sequences used in this study for amplification and sequencing were designed in the laboratory based on strain P9605 (KFDE1F4 - 5’ TGG CTC CTA CAT ATG CCA CAC GAT 3’), (KFDE2R2 - 5’ TCT GTC ACT CTG GTC TCG CTT 3’), (KFDE3F5 - 5’ CAT TGT GGC TTG TGC CAA G 3’), (KFDE4R5 - 5’ CTT GGC ACA AGC CAC AAT G 3’), (KFDE5F6 - 5’ GAA CCG CAY GCT GTG AAA ATG 3’), (KFDE6R6 -5’ CAT TTT CAC AGC RTG CGG TTC 3’), (KFDE7F6 - 5’ TAG TCA TGG AGG TGA CTT 3’), (KFDE8F5 - 5’ TGA CTA GTG GAG TGG ATC CT 3’), (KFDE9R3 - 5’ TCG CAG GTG ACA TGA CCA CTC T 3’), (KFDE107F - 5’ CAT CTA TGT TGG TGA GCT GAG 3’), (KFDE117R - 5’ CTC AGC TCA CCA ACA TAG ATG 3’), (KFDE12R4 - 5’ TGA TGA TAG CAT GCC TCC T 3’), (KFDE13R5 - 5’ TGT CAT TGT CAA CAC AAG T 3’), (KFDE14F8 - 5’ GTG GAG GCT GTG CTC AAC 3’), (KFDE15R8 - 5’ GTT GAG CAC AGC CTC CAC 3’).
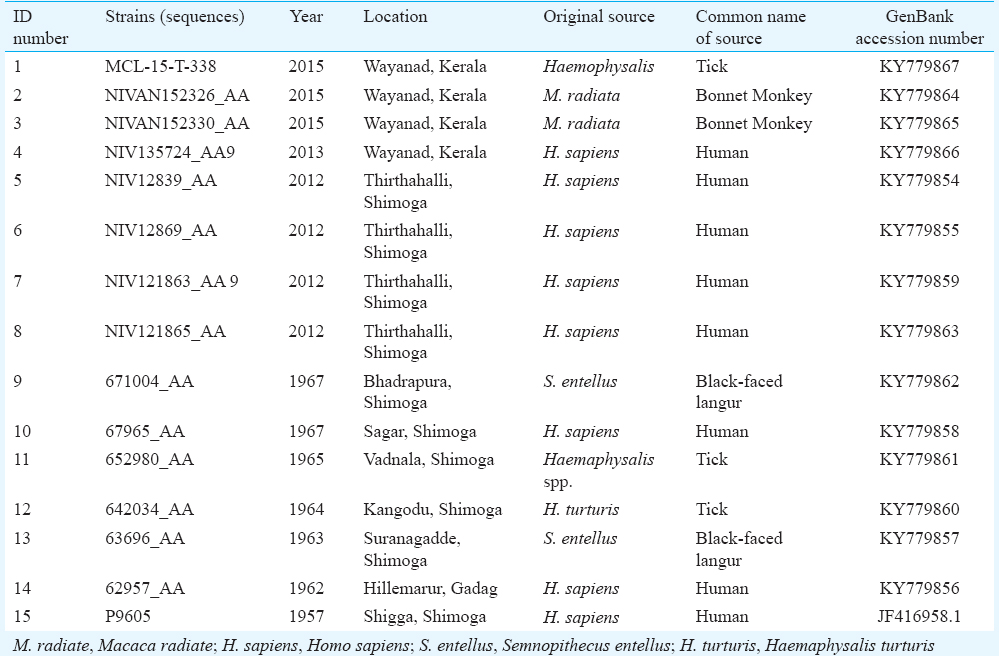
The E gene was amplified with primer set KFDE1F4 and KFDE15R8 to get polymerase chain reaction (PCR) products of 1.7 kb, which was checked in one per cent agarose gel and purified using the Qiagen Gel Extraction Kit as per the standard protocol (Qiagen). A bigger stretch than E gene was amplified and sequenced. The purified DNA was used as the target for direct nucleotide sequencing using a Big Dye Terminator Kit (Applied Biosystems, Inc., USA), followed by analysis in an ABI 3100 Automated DNA Sequencer (Applied Biosystems). All the primers as mentioned above were used for sequencing. Sequences were subjected to a Basic Local Alignment Search Tool analysis (https://blast.ncbi.nlm.nih.gov/Blast.cgi) for confirming their specificity to KFD E gene. Total number of strains used in this study was 14, and additionally, one sequence was taken from GenBank. These sequences were considered based on different geographical areas, source and year of isolation. Sequences considered in this study are enlisted in Table I along with GenBank accession number.
Phylogenetic analysis of the set of amino acid sequences was carried out using the Molecular Evolutionary Genetic Analyses (MEGA 5.0) package13. Multiple sequence alignment performed with ClustalW implementation in MEGA513 considering default parameters. The phylogenetic tree was constructed using the neighbour-joining algorithm and bootstrap (10,000 replications) was used as a test of phylogeny. Pairwise alignment of all possible pairs of sequences from the dataset was performed using the ALIGN algorithm as implemented in the ISHAN package14. In silico prediction of B-cell epitopes (antigenic determinants) was performed for each sequence of the dataset (KFD E-proteins) using the Kolaskar method15 as implemented in the B-cell epitope prediction server at Immune Epitope Database (www.immuneepitope.org/).
The 3D structures of the E-proteins from strains P9605 and MCL-15-T-338 were predicted using the SwissModel Online Workstation. The template chosen (based on the automatic template selection mode) was the E-protein of TBEV (PDB ID: 1svb). Predicted structures were evaluated by the PROCHECK analyses (https://www.ebi.ac.uk/thornton-srv/software/PROCHECK/). Visualization of all the molecular structures and rendering of images were carried out in Discovery Studio v.3.0 (Accelyrs Inc., USA). The surface electrostatics of the proteins was studied using NOC software16. Energy minimization of the modelled structures and structural comparisons were performed using the GROMOS96 force field application in Swiss PDB-Viewer (SPDBV)17. The sequences were subjected to PROSITE analyses (https://www.expasy.org/) for the prediction of functional sites.
Results
Phylogenetic analyses for the dataset were obtained with E gene and amino acids from JEV (Nakayama strain) and TBEV (strain 2517-05) as outliers. The multiple sequence analyses revealed that all the KFD amino acid sequences were highly conserved. It was observed that the sequences from Kerala outbreak of 2015, namely MCL-15-T-338 (Tick, Kerala), NIVAN152326 (Monkey, Kerala) and NIVAN152330 (Monkey, Kerala), formed a separate cluster (Figure 1) and had a few mutations. These mutations in Kerala sequences with respect to the KFD reference strain P9605 are enlisted in Table II.
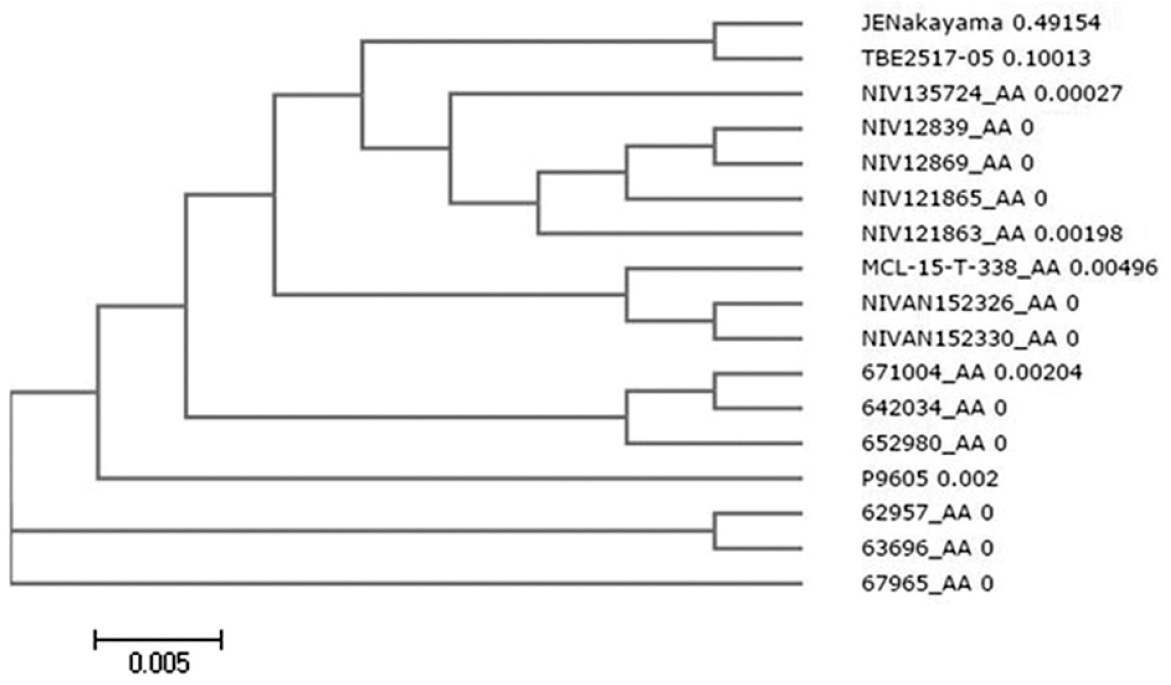
- Phylogenetic tree of Kyasanur Forest disease E-protein sequences with Japanese encephalitis virus (JEV, Nakayama strain) and tick-borne encephalitis virus (TBEV, strain 2517-05) as outlier.
The ectodomains (amino acids 1-390) of the E-protein sequences from the dataset were subjected to pairwise comparison (all possible pairs) using the ALIGN algorithm as implemented in the ISHAN package. Identity (%) of amino composition between each pair of sequences was calculated from these alignments (Table III). Minimum identity (99.2%) in amino acid composition was observed between the strains P9605 and MCL-15-T-338. This indicated that sequences from the recent outbreak deviated from the earlier ones.
The B-cell epitopes predicted based on the Kolaskar method, for all sequences were compared. These were conserved in all the sequences. It was observed that all the sequences were highly conserved and so were the epitopes. B-cell epitopes of P9605 and MCL-15-T-338 are compared (data not shown).
Since no 3D structure information for the KFDV E-protein has been reported experimentally, the structure was predicted using TBEV E-protein as template (1svb.pdb) covering 1-390 amino acids. The 3D structures of the E-proteins from the strains P9605 and MCL-15-T-338 were modelled. The identity in amino acid composition of 1svb with P9605 and MCL-15-T-338 was 81.77 and 81.4 per cent, respectively. Superposition of the P9605 and MCL-15-T-338 with respect to 1SVB.pdb generated root mean square difference (RMSD) of 0.08Å and 0.10Å, respectively, involving backbone atoms. Superposition of the 3D structures indicated that the models had identical fold of the backbone, with RMSD 0.03Å (data not shown). PROCHECK analyses for the predicted 3D structures revealed that the occupancy of Ramachandran plot was 99.7 per cent (favourable and additional favourable regions) for P9605 and 99.8 per cent for MCL-15-T-338 excluding glycine and proline residues in each case. The minimized energy of the structures for P9605 and MCL-15-T-338 was found to be −17803.0 and −17389.21 kJ/mol, respectively. These indicated that the predicted models were of good quality (data not shown). The occurrence of the mutation D239N changed the composition of epitope 239-DRLVEFG-245 in P9605 to 239-NRLVEFG-245 in MCL-15-T-338. However, this did not affect the average antigenicity of the epitope.
3D structure analyses revealed that the mutation D239N in the Kerala outbreak sequence MCL-15-T-338 occured in DII and resulted in changes in the surface contour locally and alteration in the surface electrostatics. This might affect binding or interactions with other biomolecules. The detailed analyses of functional sites using PROSITE predictions indicated that this mutation did not alter any of the functional sites. The predicted functional sites are enlisted in Table IV.
Discussion
In flaviviruses, the envelope protein (E-protein) is located on the membrane and interacts with the host immune system. Antibodies are raised against the E-protein. These antibodies are detected in the ELISA-based diagnostic kits. Hence, studying the E-protein is most important as mutations in viral membrane proteins may lead to emergence of new strain that escapes herd immunity and may lead to vaccine failure. Molecular level understanding of the process of neutralization or escape is critical for successful development and improvement of vaccines. Hence, study of the structural and functional aspects of the KFDV E-protein is vital for the understanding of virus-cell interactions as well as the biology of the virus18. The ectodomain of the Flavivirus E-protein consists of three domains each of which contains potential epitopes that can induce antibody in the host19. Although B-cell epitopes for KFDV have not been determined yet by experimentation, in our study, we concentrated on the prediction of B-cell epitopes on E-protein using bioinformatics techniques.
The predicted epitopes may be used in improving vaccine or for developing diagnostic kits in future. Further experimental studies are required for the determination of immunogenicity and protection effects of the predicted epitopes. Whether the mutations really have any effect on vaccine efficacy is a matter of future investigation, though there are no reports from the population of the affected area so far.
There were no changes in the structure of the virus that led to the spread to newer areas. The observed mutations (or amino acid differences) in the E-protein of recent strains did not alter the antigenicity and 3D structure of the envelope protein drastically. Furthermore, there are no reports of vaccine failure in the endemic areas. Hence, no correlation could be established between the amino acid differences on E-protein (antigenicity) and detection in new geographical areas. Such detection must be due to other factors such as movement of people and tourism, migration of monkeys and spread of infected vectors. However, this is a matter of investigation and the most possible reason could be that earlier no diagnosis tools were available and this disease was never considered outside of five districts of Karnataka State.
Financial support & sponsorship: None
Conflicts of Interest: None.
References
- Summary of preliminary report of investigations of the virus research centre on an epidemic disease affecting forest villagers and wild monkeys of Shimoga district, Mysore. Indian J Med Sci. 1957;11:341-2.
- [Google Scholar]
- New focus of Kyasanur forest disease virus activity in a tribal area in Kerala, India, 2014. Infect Dis Poverty. 2015;4:12.
- [Google Scholar]
- Structure-based mutational analysis of several sites in the E protein: Implications for understanding the entry mechanism of Japanese encephalitis virus. J Virol. 2015;89:5668-86.
- [Google Scholar]
- Diagnosis of Kyasanur forest disease by nested RT-PCR, real-time RT-PCR and IgM capture ELISA. J Virol Methods. 2012;186:49-54.
- [Google Scholar]
- Spread of Kyasanur forest disease, Bandipur tiger reserve, India, 2012-2013. Emerg Infect Dis. 2013;19:1540-1.
- [Google Scholar]
- Outbreak of Kyasanur forest disease in Thirthahalli, Karnataka, India, 2014. Int J Infect Dis. 2014;26:132-4.
- [Google Scholar]
- Kyasanur forest disease: An epidemiological view in India. Rev Med Virol. 2006;16:151-65.
- [Google Scholar]
- Outbreak of Kyasanur forest disease (monkey fever) in Sindhudurg, Maharashtra state, India, 2016. J Infect. 2016;72:759-61.
- [Google Scholar]
- Recent scenario of emergence of Kyasanur forest disease in India and public health importance. Curr Trop Med Rep. 2016;3:7-13.
- [Google Scholar]
- Kyasanur forest disease. In: Monath TP, ed. Arboviruses: Epidemiology and ecology. Boca Raton (FL): CRC Press; 1990. p. :93-116.
- [Google Scholar]
- Delineation of an epitope on domain I of Japanese encephalitis virus envelope glycoprotein using monoclonal antibodies. Virus Res. 2011;158:179-87.
- [Google Scholar]
- Recent ancestry of Kyasanur forest disease virus. Emerg Infect Dis. 2009;15:1431-7.
- [Google Scholar]
- MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28:2731-9.
- [Google Scholar]
- A semi-empirical method for prediction of antigenic determinants on protein antigens. FEBS Lett. 1990;276:172-4.
- [Google Scholar]
- Molecular basis of antigenic drift in influenza A/H3N2 strains (1968-2007) in the light of antigenantibody interactions. Bioinformation. 2011;6:266-70.
- [Google Scholar]
- Antigenic variability in neuraminidase protein of influenza A/H3N2 vaccine strains (1968–2009) Bioinformation. 2011;7:76.
- [Google Scholar]
- Ancient ancestry of KFDV and AHFV revealed by complete genome analyses of viruses isolated from ticks and mammalian hosts. PLoS Negl Trop Dis. 2011;5:e1352.
- [Google Scholar]
- A ligand-binding pocket in the dengue virus 2 envelope glycoprotein. Proc Natl Acad Sci U S A. 2003;100:6986.
- [Google Scholar]






