Translate this page into:
Bacterial aetiology of neonatal meningitis: A study from north-east India
*For correspondence: jmahanta@gmail.com
-
Received: ,
This is an open access article distributed under the terms of the Creative Commons Attribution-NonCommercial-ShareAlike 3.0 License, which allows others to remix, tweak, and build upon the work non-commercially, as long as the author is credited and the new creations are licensed under the identical terms.
This article was originally published by Medknow Publications & Media Pvt Ltd and was migrated to Scientific Scholar after the change of Publisher.
Sir,
Neonatal infection is one of the major causes of neonatal deaths in India1. Bacterial meningitis being a life-threatening condition requires prompt diagnosis and treatment, but diagnosis in newborn is a challenge for clinicians because symptoms and signs are often subtle and non-specific at its early stage2. It has been reported that neonatal meningitis can occur in the absence of bacteraemia and no single cerebrospinal fluid (CSF) value can exclude its presence3. Several studies from India have reported the spectrum of bacterial pathogens and the existence of antibiotic resistance among the isolates from neonatal sepsis/meningitis cases4567. We have earlier isolated rarely encountered bacteria including Neisseria meningitidis serogroup Y8, Sneathia9 and Globicatella (unpublished) species from CSF of neonates from north-east India. The present study was done with an aim to find out the profile of bacterial pathogens causing neonatal meningitis in this part of the country. Since in most cases antimicrobial treatment was initiated before performing lumbar puncture, the study was also aimed to find out if 16S rRNA polymerase chain reaction (PCR) had a role in identifying organism and meningitis in CSF culture-negative neonates with sepsis.
This was a hospital-based study carried out for two consecutive years (January 2013-January 2015) in the Neonatal Unit of Assam Medical College and Hospital, a tertiary care hospital at Dibrugarh, Assam, India. About 2000-3000 inborn (IB) neonates require admission in the unit per year. Neonatal mortality is 20 per 1000 live births among the IB cases. CSF samples were obtained from neonates when there was clinical sepsis/meningitis or proven sepsis and in whom blood culture grew microorganism. The clinical criteria for suspecting meningitis included late-onset sepsis with or without seizure, bulging anterior fontanelle, vacant stare, high-pitched cry, neck retraction. In case of early-onset sepsis, meningitis was looked for in all neonates with culture-proven or clinical sepsis. The microbiological investigations of the CSF samples were done at the Regional Medical Research Centre, Dibrugarh. The CSF cytological and biochemical tests were done at the ICMR-Regional Medical Research Centre, Dibrugarh. Sepsis screen and blood culture reports, if available, were noted down from the patient record sheet. The demographic and clinical data of the neonates were recorded. Early and late onset of sepsis were defined as infection within the first 72 h of life and >72 h of life, respectively.
CSF samples were processed for microbiological investigation within one hour of collection. Briefly, the CSF was centrifuged at 1000 × g for 10-15 min. One drop of sediment was used to prepare the Gram stain and one drop was used to streak the primary culture medium (blood agar, chocolate agar and MacConkey agar). The inoculated plates were incubated under aerobic conditions at 37°C with 5 per cent CO2. Conventional identification methods based on characteristics such as morphology, appearance in culture medium and biochemical tests were used to identify the isolated organisms. When results were inconclusive diagnosis was made using PCR targeting genes specific for different bacteria including Acinetobacter baumannii10, Enterococcus11, Escherichia coli12 and N. meningitidis13. The DNA was extracted using PureLink Genomic DNA Kit (Invitrogen, USA) following the manufacturer's instructions. Bacterial isolates obtained on culture were also reconfirmed by 16S rRNA sequencing using universal primers, forward primer: 5’-AGA GTT TGA TCC TGG CTC AG-3’ and reverse primer: 5’-ACG GCT ACC TTG TTA CGA CTT-3’ (Promega, Madison, USA) using the thermal profile : 4 min of initial denaturation at 94°C, followed by 35 cycles of 1 min denaturation at 94°C, 1 min primer annealing at 58°C, and 2 min elongation at 72°C, and finally extension for 7 min at 72°C. The supernatant was used for antigen detection by latex agglutination test, following the manufacturer's instructions (Directigen Meningitis Combo Test, BD, USA). Antigen detection was performed when Gram stain of CSF was inconclusive despite a suggestive cytology and clinical findings suggestive of meningitis. It could not be done in situations when the quantity of CSF received was insufficient or if the CSF was haemorrhagic.
I6S rRNA gene amplification was also carried out directly on CSF sample to exclude the presence of any organism for the culture-negative samples. For this, DNA was extracted by boiling 200 μl of the available CSF sample for 10 min, followed by centrifugation at 12,000 × g for 10 min. The supernatant was used as DNA template. The amplified product was purified using a commercial kit (High Pure PCR Product Purification Kit, Version 15, Roche, Germany) as per the manufacturer's instructions. Assembly, alignment and consensus of the DNA sequences were made using CodonCode Aligner version 2.0 (http://www.codoncode.com/aligner/download.htm). A BLAST search (https://blast.ncbi.nlm.nih.gov/Blast.cgi) was made to compare our query sequence with NCBI database of sequences.
The Kirby-Bauer disc diffusion method14 was used for antimicrobial susceptibility testing of the CSF isolates. The guidelines of the Clinical and Laboratory Standards Institute (CLSI)14 were used for interpretation as resistant, intermediate sensitive and sensitive. The following antibiotics were used: ampicillin (AMP) 10 μg; ampicillin/sulbactam (A/S) 10/10 μg; amikacin (AK) 30 μg; cefotaxime (CTX) 30 μg; ciprofloxacin (CIP) 5 μg; gentamicin (GEN) 10 μg; linezolid (LZD) 30 μg; netilmicin (NET) 30 μg; piperacillin (PI) 100 μg; piperacillin/tazobactam (P/T) 100/10 μg; vancomycin (VA) 30 μg. VA-resistant Enterococcus (VRE) and coagulase-negative Staphylococcus (CoNS) isolates were confirmed by minimum inhibitory concentration (MIC) using E-test strip14. LZD resistance in Enterococcus isolates was based on zone size interpretation as per the CLSI interpretative criteria14. Positive reports were made immediately known to the neonatologist. Data were entered into Statistical Package for Social Science version 13.0 (SPSS Inc., Chicago, USA) for risk factor analysis. Ethical clearance was obtained from Institutional Ethics Committee of Assam Medical College & Hospital as well as Regional Medical Research Centre, Dibrugarh. Written informed consent was obtained from either parent.
Of the 303 CSF samples tested, 67 were positive for pathogens. Of these, 52 were positive by culture and another 15 by 16S rRNA gene PCR. Five of these were excluded as probable contaminant. Those excluded from analysis were four isolates of Micrococcus luteus and one isolate of Kocuria spp. These were excluded as CSF parameters were not suggestive of meningitis. In two cases positive for M. luteus, blood culture report was positive but the organisms isolated were different. Repeat CSF culture could not be done in such cases.
Direct Gram stain of CSF was positive in 30 samples, of which 21 were culture positive. Direct antigen testing was positive in two samples [group B streptococci (GBS): n=1; N. meningitidis, n=1]. Those positive by either culture or 16S rRNA PCR were associated with clinical signs of meningitis, CSF parameters, sepsis screen profile and blood culture, and all of these were positive for at least one criterion.
Among bacterial isolates detected, Gram-negative bacteria were predominant (n=32, 48%) as compared to Gram-positive (n=26, 39%). Yeast was isolated on four occasions and five isolates when compared with NCBI database after sequencing of its 16S rRNA gene sequences were comparable with those of uncultured bacterium. The most frequent Gram-negative bacteria detected were A. baumannii (n=12, 18%), Klebsiella (n=8, 12%), Pseudomonas (n=6, 9%). Less frequently detected were N. meningitidis (n=2, 3%), and one each for E. coli, Sneathia, Cronobacter sakazakii, and Roseomonas cervicalis. Among the Gram-positive bacteria, Enterococcus spp. (n=10, 15%) and CoNS (n=11, 16%) were most commonly detected (Table I). E. faecium (n=7) was the most common species within the genus Enterococcus. Those detected by direct CSF 16S rRNA PCR and sequencing were Staphylococcus spp. (n=1), Staphylococcus sciuri (n=1), Klebsiella spp. (n=1), Klebsiella pneumoniae (n=1), Sneathia (n=1), Pseudomonas (n=1), GBS (n=1), A. baumannii (n=3) and uncultured bacteria (n=5).
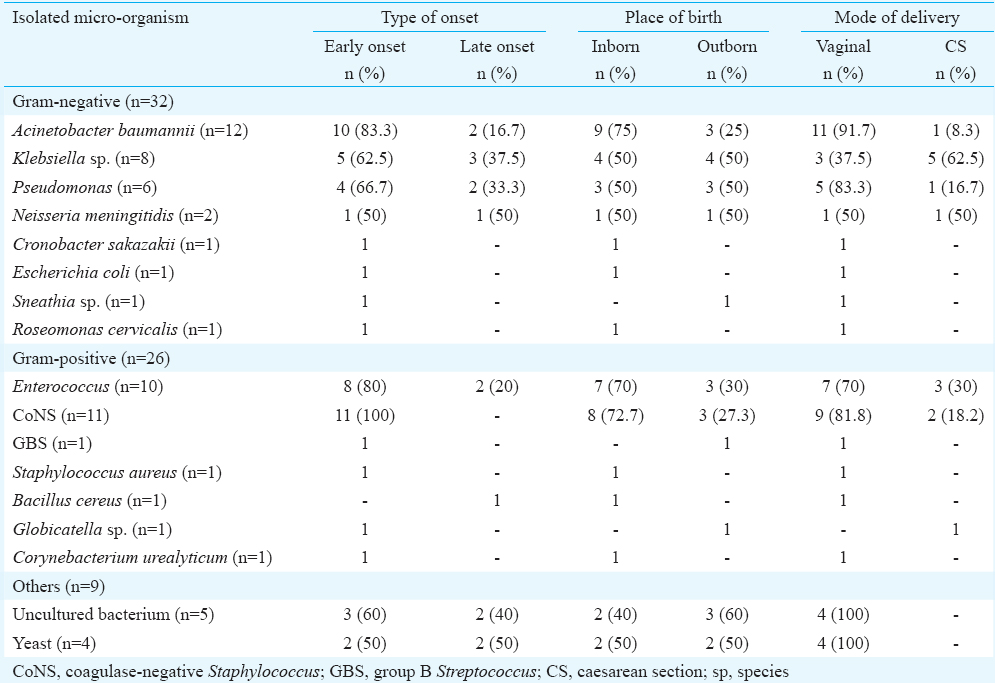
Bacillus cereus and Corynebacterium urealyticum were included as causative organisms as in these cases; blood culture was positive and C-reactive protein (CRP) was raised. In case of C. sakazakii and R. cervicalis, the CRP was raised; hence, both were also included. Sequences of a few isolates were deposited in the GenBank. The bacterial isolates were A. baumannii (KP943717, KP943719, KP943724, KP943727), K. pneumoniae (KP943718, KP943729), Pseudomonas (KP943725, KP943732), E. faecium (KP943720, KP943722), S sciuri (KP943721) and B. cereus (KP943723).
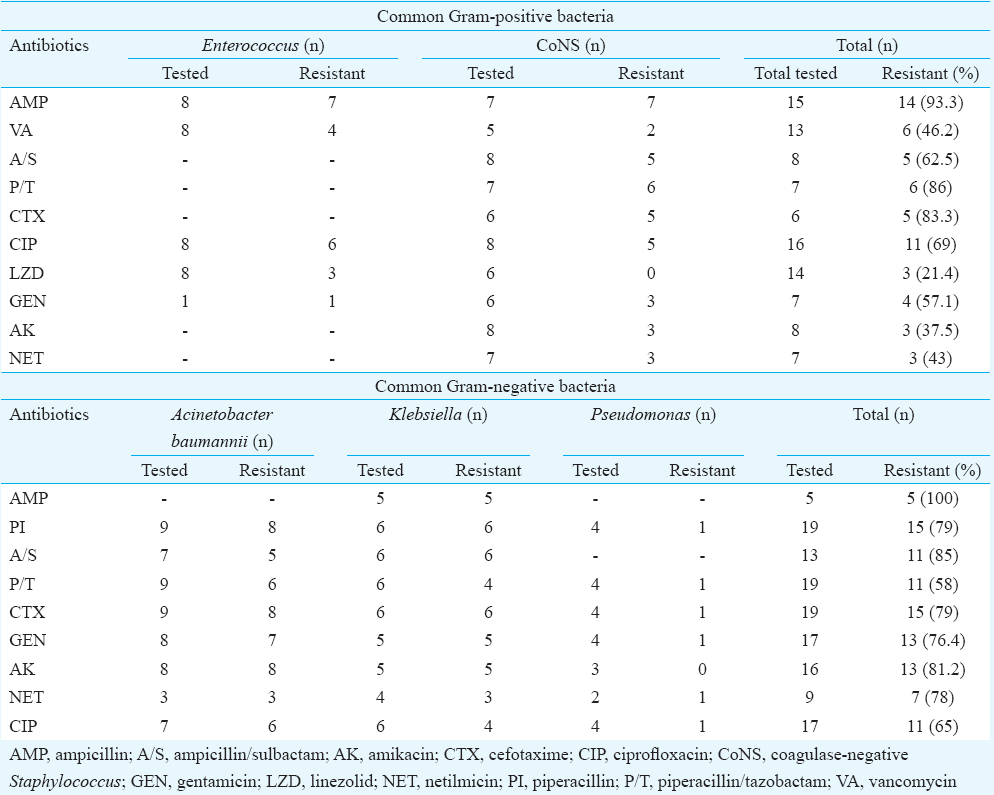
Table II shows the susceptibility pattern of the common isolates. About 63 per cent of the Gram-positive isolates were sensitive to AK, 57 per cent to NET, 38 per cent to A/S, 31 per cent to CIP, 14 per cent to P/T and 6.7 per cent to AMP. VA and LZD sensitivity was seen in 54 and 79 per cent, respectively. About 42, 35, 22 and 19 per cent of the Gram-negative isolates were sensitive to P/T, CIP, NET and AK, respectively. AMP resistance was seen in 100 per cent of the Gram-negative isolates tested. About 83 per cent of the Gram-positive and 79 per cent of the Gram-negative isolates tested were resistant to CTX. Sepsis screen result showed raised CRP in 52.2 per cent (35/67). Clinical evaluation of those neonates who had a pathogen detected in its CSF sample showed seizure in 47.8 per cent (n=32), lethargy in 32.8 percent (n=22), bulging anterior fontanelle in 12 per cent (n=8), high-pitched cry in 4.5 per cent (n=3), neck retraction in 3 per cent (n=2), sub-galeal bleed in 1.5 per cent (n=1) and vacant stare in 9 per cent (n=6).
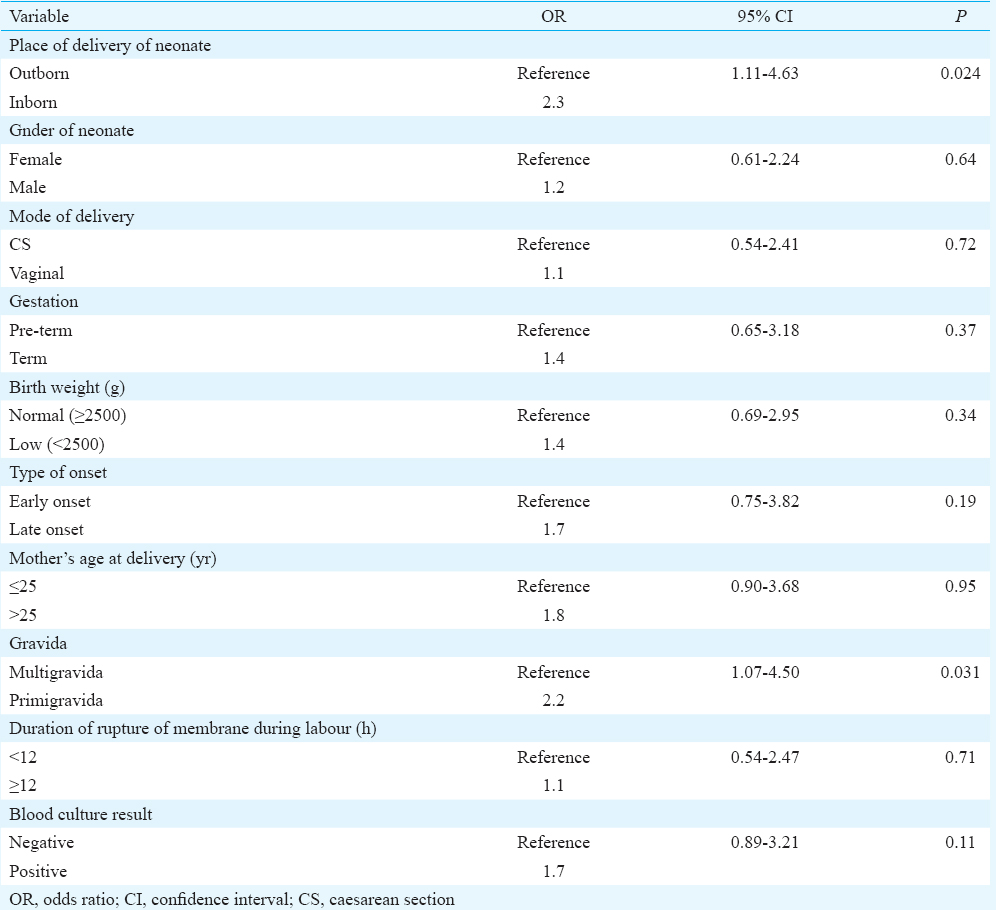
Blood culture was positive in 23 of the 67 neonates whose CSF samples were microbiologically positive. Thus, concordance between CSF positivity and blood culture positivity was seen in 34.3 per cent cases. Twenty one (6.9%) neonates expired of whom three had a positive CSF yield. Multivariate logistic regression analysis for risk factors revealed that two variables, namely, primigravida (OR: 2.2; CI: 1.07-4.50; P=0.031) and IB neonates (OR: 2.3; CI: 1.11-4.63; P=0.024), were found to be independent predictors for meningitis. Regression was adjusted for different variables including gender, mode of delivery, gestation, birth weight, time of onset of symptoms, blood culture positivity, maternal age and duration of rupture of membrane (Table III).
The most common symptom among neonates was seizure and lethargy. Similar observation has been made earlier from India6. In the present study, only 12 per cent cases showed increased CSF protein and 7.5 per cent showed decreased glucose. Normal CSF parameters in the presence of meningitis have been described by others3. In our study, 68.6 per cent neonates with microbiologically proven meningitis were males; and 54 per cent were of low birth weight (LBW), 11 per cent were very LBW and 28 per cent were pre-term. Kaul et al6 have also observed high frequency of meningitis and sepsis among LBW neonates. A. baumannii, Klebsiella, CoNS and Enterococcus were common bacterial isolates in our study similar to other studies from India415. A study on neonatal meningitis from south India found Streptococcus pneumoniae to be the most common pathogen followed by Haemophilus influenzae and Pseudomonas in neonates5. Pseudomonas and Klebsiella have also been reported as predominant isolates in neonates with sepsis7. B. cereus and C. sakazakii, which are environmental isolates, have also been reported as a cause for neonatal sepsis and meningitis1617. Thus, continued surveillance of these agents is requisite for deciding empirical treatment as prompt treatment is important in such cases. Rare pathogens were also isolated in the present study. Two of them including N. meningitidis serogroup Y and Sneathia have been described elsewhere89.
Only 15 of the 67 samples positive for pathogens were found positive using PCR. In the present study, all direct CSF PCR-positive samples were found to relate either clinically or with CSF parameters for meningitis or associated with a positive blood culture. Gram-negative bacteria were predominantly isolated as compared to Gram-positive bacteria as has been reported in other Indian studies also47.
Early-onset bacterial neonatal infection is known to be caused by those pathogens colonizing or infecting the maternal genital tract or in the delivery area18. In the present study, Enterococcus has been seen to be isolated on a higher percentage from those who had been delivered vaginally and presented with early onset of symptoms. E. coli, GBS, Globicatella, Sneathia all known as vaginal colonizers, have been isolated from those who were born through vaginal delivery and had an early onset of symptoms. A. baumannii, C. sakazakii, CoNS, Pseudomonas spp., Roseomonas and Bacillus can be isolated from various environmental sources including the hospital environment. Thus, it is likely that these can colonize the neonate and cause early-onset infection.
Even though resistance to the commonly used antibiotics in neonates was seen in the present study, the neonates responded clinically following treatment. Observation of low sensitivity to commonly used antibiotics has been observed in other studies also47. In our study 4.5 per cent neonates expired. Low mortality observed at this tertiary care centre could be due to the prompt treatment as a result of appropriate and timely diagnosis. Further, 22.4 per cent culture negative samples were found positive by PCR implying the importance of PCR in such cases. The negative culture could be due to prior antibiotic use. Importance of PCR in meningitis cases has been shown earlier as the results are not affected by prior antibiotic use19. Thus, continuous monitoring of causative agents of neonatal infection, control of emergence of drug resistance among them through specific and appropriate duration of treatment, preventive intervention for transmission to the neonate can be an appropriate approach for controlling neonatal deaths.
Acknowledgment
The study was supported by an extramural grant received from the Indian Council of Medical Research (ICMR), New Delhi.
Conflicts of Interest: None.
References
- Causes of neonatal and child mortality in India: A nationally representative mortality survey. Lancet. 2010;376:1853-60.
- [Google Scholar]
- Diagnosis and treatment of bacterial meningitis in the newborn. Niger J Paediatr. 2013;40:6-14.
- [Google Scholar]
- Neonatal meningitis: What is the correlation among cerebrospinal fluid cultures, blood cultures, and cerebrospinal fluid parameters? Pediatrics. 2006;117:1094-100.
- [Google Scholar]
- Profile of neonatal septicaemia at a district-level sick newborn care unit. J Health Popul Nutr. 2012;30:41-8.
- [Google Scholar]
- Bacteriological profile of community acquired acute bacterial meningitis: A ten-year retrospective study in a tertiary neurocare centre in South India. Indian J Med Microbiol. 2007;25:108-14.
- [Google Scholar]
- Importance of obtaining lumbar puncture in neonates with late onset septicemia a hospital based observational study from North-west India. J Clin Neonatol. 2013;2:83-7.
- [Google Scholar]
- Bacterial isolates of early-onset neonatal sepsis and their antibiotic susceptibility pattern between 1998 and 2004: An audit from a center in India. Ital J Pediatr. 2011;37:32.
- [Google Scholar]
- Neonatal meningitis due to Neisseria meningitidis serogroup Y. Indian Pediatr. 2014;51:757.
- [Google Scholar]
- Sneathia species in a case of neonatal meningitis from Northeast India. Oxf Med Case Reports. 2014;2014:112-4.
- [Google Scholar]
- Development of a multilocus sequence typing scheme for characterization of clinical isolates of Acinetobacter baumannii. J Clin Microbiol. 2005;43:4382-90.
- [Google Scholar]
- Development of a PCR assay for rapid detection of enterococci. J Clin Microbiol. 1999;37:3497-503.
- [Google Scholar]
- Escherichia coli MLST Database. Available from: http://www.mlst.warwick.ac.uk/mlst/dbs/Ecoli/
- Simultaneous approach for nonculture PCR-based identification and serogroup prediction of Neisseria meningitidis. J Clin Microbiol. 2000;38:855-7.
- [Google Scholar]
- Clinical and Laboratory Standards Institute. Performance standards for antimicrobial susceptibility testing. 20th informational supplement. Wayne, PA: Clinical and Laboratory Standards Institute; 2010.
- Multi-drug-resistant, non-fermenting, Gram-negative bacilli in neonatal sepsis in Kolkata, India: A 4-year study. Paediatr Int Child Health. 2014;34:56-9.
- [Google Scholar]
- Bacillus cereus bloodstream infection in a preterm neonate complicated by late meningitis. Case Rep Infect Dis 2012 2012:358789.
- [Google Scholar]
- Enterobacter sakazakii infections among neonates, infants, children, and adults. Case reports and a review of the literature. Medicine (Baltimore). 2001;80:113-22.
- [Google Scholar]
- How can the microbiologist help in diagnosing neonatal sepsis? Int J Pediatr 2012 2012:120139.
- [Google Scholar]
- Application of 16S rDNA based seminested PCR for diagnosis of acute bacterial meningitis. Indian J Med Res. 2009;129:182-8.
- [Google Scholar]





