Translate this page into:
Attenuation of quorum sensing-regulated behaviour by Tinospora cordifolia extract & identification of its active constituents
Reprint requests: Dr Krutika B. Desai, Department of Microbiology, Mithibai College, Bhakti Vedanta Swami Marg Vile Parle (West), Mumbai 400 056, Maharashtra, India e-mail: krutika.desai@mithibai.ac.in
-
Received: ,
This is an open access article distributed under the terms of the Creative Commons Attribution-NonCommercial-ShareAlike 3.0 License, which allows others to remix, tweak, and build upon the work non-commercially, as long as the author is credited and the new creations are licensed under the identical terms.
This article was originally published by Medknow Publications & Media Pvt Ltd and was migrated to Scientific Scholar after the change of Publisher.
Abstract
Background & objectives:
The pathogenicity of the nosocomial pathogens, Pseudomonas aeruginosa and Acinetobacter baumannii is regulated by their quorum sensing (QS) systems. The objective of the present study was to examine the effect of the cold ethyl acetate extract of Tinospora cordifolia stem on virulence and biofilm development in the wild type and clinical strains of P. aeruginosa and A. baumannii. The study was further aimed to identify the probable active constituents in the plant extract.
Methods:
P. aeruginosa virulence factors viz., LasA protease, LasB elastase and pyocyanin production were analyzed spectrophotometrically. Biofilm formation was studied using crystal violet staining-microtitre plate assay. The plant extract was fractionated using silica gel column chromatography and the most active fraction was derivatized using silylation and analyzed by gas chromatography-mass spectrometry (GC-MS). In silico testing of the molecules identified in GC-MS was performed, for binding to the P. aeruginosa LasI and LasR proteins, to predict the QS inhibitory molecules.
Results:
The plant extract inhibited three major virulence factors in P. aeruginosa; it exhibited enhanced biofilm formation in P. aeruginosa while decreased biofilm development in A. baumannii. The most active fraction obtained from column chromatography, exhibited suppression of virulence as well as biofilm in both the organisms. Docking scores were calculated for all the molecules identified in GC-MS, and high docking scores were obtained for 2,3,4-triacetyloxybutyl acetate, methyl 16-methyl heptadecanoate, 2-(5-ethenyl-5-methyloxolan-2-yl)propan-2-ol, methyl hexadecanoate and 2-methoxy-4-vinyl phenol.
Interpretation & conclusions:
The compounds showing high docking scores could probably be the QS inhibitors. These molecules can be screened further for the development of new anti-infective drugs.
Keywords
Acinetobacter baumannii
Biofilm
LasI
LasR
Pseudomonas aeruginosa
quorum sensing
Tinospora cordifolia
virulence
Targeting virulence of the pathogen is considered a promising antimicrobial approach, wherein drugs act by depriving microorganisms of their virulence properties, protect further tissue damage or spread of infection, which finally results in the pathogen's clearance by the host immune system1. The Gram negative bacterial pathogens, Pseudomonas aeruginosa and Acinetobacter baumannii have been commonly implicated in healthcare associated infections23. P. aeruginosa infections involve several cell-associated (pilus, nonpilus adhesins, lipopolysaccharide, biofilm) and extracellular (Las A and Las B proteases, haemolysins, exotoxin A, exoenzyme S, pyocyanin) virulence factors4. The mechanisms of A. baumannii infections are not clear; however, certain virulence factors have been marked, such as pilus, lipopolysaccharide, OmpA, and biofilm5. The production of extracellular virulence factors, and biofilm in P. aeruginosa as well as biofilm development in A. baumannii is under the regulation of quorum sensing (QS)46. QS is a cell density dependent mechanism, wherein a threshold concentration of small signals called as autoinducers results in the activation of a transcriptional regulator protein. This activated protein regulates the expression of genes, which control coordinated group behaviours in bacteria. Acyl homoserine lactones (Acyl-HSLs or AHLs) are commonly produced as autoinducers by many Gram-negative bacteria1. P. aeruginosa produces a long acyl chain-HSL, 3-oxo-C12-HSL, as well as a short acyl chain-HSL, C4-HSL, which regulate the production of virulence factors as well as auto-induction4. In A. baumannii, a long acyl chain-HSL, 3-hydroxy-C12-HSL is known to regulate biofilm maturation6. Inhibition of QS may help to restrain virulence and biofilm development during infections, thus controlling the associated tissue damage and infection spread, leading to bacterial clearance by the host immunity. It may also sensitize the organism to antibiotics, by inhibiting biofilm development.
A preliminary screening study for anti-QS activity of Tinospora cordifolia revealed significant anti-QS potential in the ethyl acetate extracts of the dried stem of this plant7. In the present study, cold ethyl acetate extract (CEE) of this plant was explored further for its activity against QS dependent virulence and biofilm development in P. aeruginosa and A. baumannii. The effect of plant extract was studied on three major virulence factors of P. aeruginosa, LasA protease, LasB elastase and pyocyanin as also on biofilm development in P. aeruginosa and A. baumannii. The study was extended further to purify the crude extract and identify its active components using column chomatography, gas chromatography-mass spectrometry (GC-MS) and in silico molecular docking.
Material & Methods
All media and medium components were procured from Hi-Media Laboratories, Mumbai. All organic solvents and other chemicals were purchased from Qualigens Fine Chemicals, India.
Bacterial strains and culture conditions: Pseudomonas aeruginosa PAO1 and Acinetobacter baumannii MTCC1425 were used as reference strains. Escherichia coli MG4/pKDT17 and Chomobacterium violaceum ATCC12472 were used as AHL reporter strain. Staphylococcus aureus MTCC737 strain was used for LasA protease assay. Luria Bertani (LB) medium was used for the bacterial growth. E. coli MG4/pKDT17 was grown in LB medium containing 100 μg ampicillin.
Tests and controls: CEE dissolved in 100 per cent dimethyl sulphoxide (DMSO), was used as the test in all assays. Azithromycin dihydrate (Sisco Research Laboratories, India), dissolved in sterile phosphate buffered saline (PBS), pH 6.0, was used as positive control for QS inhibition, and inhibition of QS regulated virulence and biofilm development8. Azithromycin was used at a standardized concentration of 4.0 μg/ml. LB broth, with or without plant extract, was used as blank. DMSO and PBS were maintained as negative controls. Same volumes of the test and controls were used in all the assays.
Plant material collection and authentication: Dried stem of Tinospora cordifolia (Willd.) Miers was purchased from local vendor, authenticated and voucher deposited (Number: S/B-122) at Agharkar Research Institute, Pune, Maharashtra, India.
Preparation of CEE: Dried and powdered, stem (100 g) was macerated in 500 ml of ethyl acetate at ambient temperature (27°C) with intermittent shaking for 48 h. The extract was filtered using Whatman filter paper no. 1, dried in hot air oven at 45°C, and stored at 4°C in amber bottle.
Anti-QS activity at different concentrations: C. violaceum pigment production assay and long acyl-HSL production assay7 were performed to determine the anti-QS activity of plant extract (CEE) at the concentration range of 0.2 to 10.0 mg/ml (0.2, 0.5, 1, 2, 4, 6, 8, 10). Further, IC50 was calculated using regression analysis. The IC50 value was used as a basis to decide the test concentration for further assays.
C. violaceum pigment production assay: C. violaceum produces a short acyl-HSL (C6-HSL) which regulates production of dark purple pigment, violacein. Thus, inhibition of short acyl-HSL in C. violaceum is indicated by inhibition of violacein production. Agar well diffusion assay was performed to evaluate inhibition of short acyl-HSLs by CEE7.
Long acyl-HSL production assay: The ability of CEE to inhibit long acyl-HSLs in P. aeruginosa and A. baumannii was tested using the reporter strain, E. coli MG4/pKDT177. This strain expresses β-galactosidase gene in response to the exogenous addition of long acyl-HSLs. The enzyme activity thus corresponds to the amount of long acyl-HSLs added, and decrease in enzyme activity indicates long acyl-HSL inhibition. Overnight bacterial cultures of P. aeruginosa and A. baumannii were diluted with fresh LB broth, up to an OD600 of 0.2. Then, 0.1 ml of CEE (final concentration of 0.2 to 10.0 mg/ml)/controls was added to 4.9 ml of the diluted culture and incubated at 37 °C for
24 h. Acyl-HSL obtained in the culture supernatant was extracted with ethyl acetate, twice; ethyl acetate layers were pooled and evaporated to dryness under nitrogen. The dried extract was reconstituted in LB broth and added to 2.0 ml of overnight grown reporter strain culture and incubated for five hours at 37° C to induce the expression of β-galactosidase. The enzyme activity was determined as described by Miller using ortho-nitrophenyl-ß-galactopyranoside (ONPG) as substrate9 and expressed in terms of Miller units.
Collection of clinical isolates: Clinical isolates of P. aeruginosa and A. baumannii, obtained from wound infections, respiratory infections, urinary tract infections and bloodstream infections, were obtained from local government hospitals. The isolates were maintained as 20 per cent glycerol stocks at - 80°C.
Qualitative AHL detection: A modified protocol of cross feeding assay10 was followed. The bacterial isolates were cross streaked in concentric circles rather than parallel straight lines. The test strain (clinical isolates of P. aeruginosa and A. baumannii) was spot inoculated and the reporter strain (E. coli MG4/pKDT17) was streaked around it, one cm apart, on LB agar, previously spread with 40 μl of 20 mg/ml X-gal solution (Sigma-Aldrich, USA). Reference strains were used as positive control, and reporter strain as negative control. Blue colouration in the growth of reporter strain suggested AHL production.
Qualitative protease detection: Clinical isolates of P. aeruginosa were spot inoculated on skim milk agar plates and incubated at 37°C. After overnight incubation, halo around growth indicated protease activity. Reference strain (PAO1) was used as positive control.
Typing of clinical isolates of P. aeruginosa: The procedure of pyocin typing given by Gillies and Govan11 was modified. Due to unavailability of the Wahba indicator strains, the clinical isolates were cross streaked among themselves to check for growth inhibition of each isolate by the remaining isolates. Thus, depending on the growth inhibition pattern, the isolates were grouped.
Effect of CEE on virulence of P. aeruginosa: CEE, dissolved in DMSO, was used at a final concentration of 5 mg/ml (rounded off IC50 value) to evaluate its activity against the production of extracellular virulence factors in reference (PAO1) and clinical isolates (K04 and C03) of P. aeruginosa. The clinical strains, K04 and C03, formed the representatives for group I and group II isolates (classified on the basis of pyocin typing), respectively. Reduced enzyme activity or pyocyanin concentration in comparison to negative control suggested virulence inhibition. Per cent inhibition was calculated as [(enzyme activity/pyocyanin concentration for negative control - enzyme activity/pyocyanin concentration for CEE/azithromycin)×100] ÷ [enzyme activity/pyocyanin concentration for negative control].
Las A staphylolytic protease assay: The method of Kessler et al12 was followed, with modifications. The reaction volume was maintained at 2.0 ml instead of 1.0 ml, and the reaction temperature was optimized at 37 °C. P. aeruginosa was inoculated in 5 ml LB broth containing controls/CEE, up to an OD600 value of 0.2, and incubated for 24 h, at 37°C. Culture was centrifuged at 8,000×g, 4°C. 0.5 ml of culture supernatant was mixed with 1.5 ml of boiled (15 min) S. aureus suspension (OD600 value of ~0.6) in 0.02 M Tris-Cl buffer (pH 8.5). OD600 was measured at 0 and 3 h post incubation at 37°C, using Perkin-Elmer UV-Vis Spectrophotometer Lambda 25 model. Enzyme activity was expressed as units/ml; 1 unit corresponding to decrease in OD by 0.01.
Las B elastase assay: Enzyme activity was studied as per the method of Pearson et al13 with modifications. The bacterial cultures were grown in LB broth instead of peptone tryptic soy broth (PTSB), the substrate concentration was decreased from 20 to 10 mg Elastin-Congo Red (ECR) per ml of buffer and the incubation time was optimized to 24 h. P. aeruginosa was inoculated in 5 ml LB broth containing controls/CEE, up to an OD600 value of 0.2, and incubated for 24 h, at 37°C. Culture was centrifuged at 8,000×g, 4°C. One ml of buffer (Tris-Cl 0.1 M, CaCl2 1 mM, pH 7.5) containing 10 mg ECR substrate (Sigma-Aldrich, USA) was mixed with 0.5 ml of culture supernatant, incubated at 37°C, 24 h. The tubes were centrifuged at 8,000×g, 4°C and OD495 of the supernatant was measured. Enzyme activity was expressed as units/ml, where 1 unit represents increase in OD by 0.01.
Pyocyanin production assay: The procedure given by Huerta et al14 was followed, with modifications. The bacterial strains were grown in King's medium B for 48 h instead of culturing in LB medium with 24 h incubation. P. aeruginosa was inoculated in King's medium B broth containing controls/CEE up to OD600 value of 0.2 and incubated for 48 h, at 37°C. Pyocyanin was extracted from the culture supernatant using chloroform and measured spectrophotometrically at 690 nm. Pyocyanin concentration was calculated as mg/ml using the formula, OD690 ÷ 1615.
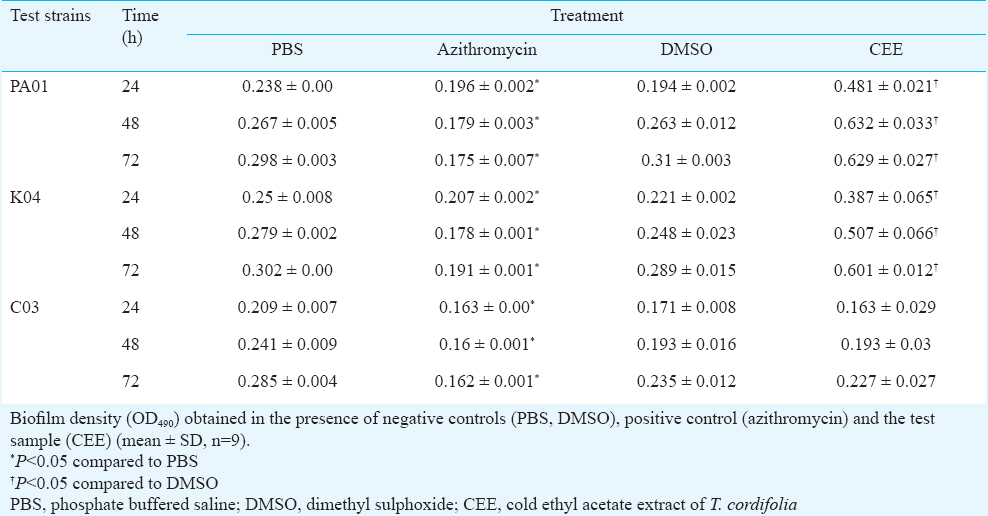
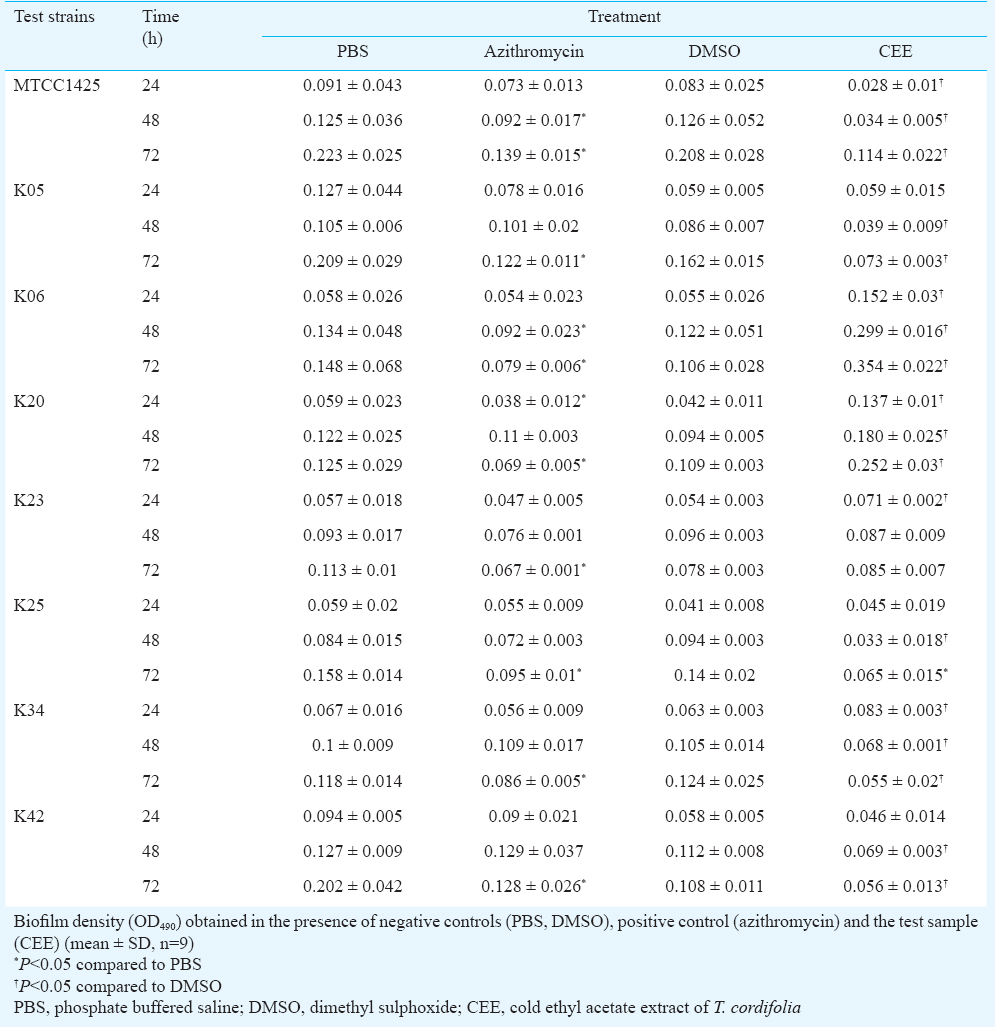
Effect of CEE on biofilm formation of P. aeruginosa and A. baumannii: Biofilm formation of the reference strains and clinical isolates of the organisms was evaluated using 96-well microtiter plate assay, as per the method of Mathur et al16, with modifications. LB broth supplemented with glucose was used for biofilm growth, biofilm fixation was done using methanol, and the bound dye was dissolved in ethanol before measuring its absorbance at 490 nm. An aliquots (200 μl) of overnight cultures diluted in LB broth (supplemented with 0.5% glucose) was added to the microtiter wells. CEE, dissolved in DMSO, was added at the concentration of 5.0 mg/ml. For positive control, azithromycin dihydrate, dissolved in PBS, pH 6.0, was used at a 4 μg/ml concentration and DMSO/PBS was used as negative control. The plates were incubated for 24, 48 and 72 h, at 37 °C. Following incubation, the contents in the wells were removed, washed with distilled water, and the adherent biofilm was fixed with methanol for 15 min. This was followed by staining with 1 per cent crystal violet dye for 15 min. Excess stain was removed, the wells were washed with distilled water and the remaining bound dye was resolubilized in 95 per cent ethanol for five min. OD490 was measured using BioRad Microplate reader Model 680 (Hercules, CA, USA). Per cent inhibition was calculated as [(biofilm density for negative control - biofilm density for CEE/azithromycin)×100] ÷ [biofilm density for negative control].
Phytochemical class detection of CEE: Standard chemical tests17 were performed for detection of carbohydrates, proteins, alkaloids, tannins, flavonoids, cardiac glycosides, anthroquinone glycosides, coumarin glycosides, saponin glycosides and steroids.
Activity guided fractionation of CEE: CEE (5 g) was subjected to silica gel (60-120 mesh) (Merck Co., India) column chromatography. A glass column of 600 mm length, 35 mm internal diameter and volume capacity of 600 ml, was packed with a silica bed of about 270 mm length. CEE was separately adsorbed on silica gel, dried, and loaded over the silica bed. Gradient elution was carried out using 100 per cent petroleum ether, followed by increasing concentration of ethyl acetate, up to 100 per cent ethyl acetate, and then increasing polarity with methanol, upto 80 per cent methanol. About 50 ml fraction volume was collected, and based on thin layer chromatography (TLC) profile, the fractions were pooled. In total, 11 fractions were obtained and further evaluated for anti-QS, anti-virulence, anti-biofilm and anti-bacterial activities. The fraction showing significant (P<0.05) anti-QS, anti-virulence, anti-biofilm activities and absence of antibacterial activity, was subjected to GC-MS analysis.
Identification of components in active fraction: One mg of the fraction (fraction 5) was derivatized by silylation reaction using 100 μl N-methyl-trimethylsilyltrifluoroacetamide (MSTFA) (Sigma-Aldrich, USA) and 50 μl pyridine, at 60°C, for 30 min. The mixture was dried, redissolved in ethyl acetate and analyzed using GC-MS18. Shimadzu GCMS-QP2010 ultra equipped with a quadrapole mass spectrometer (Tokyo, Japan) was used. Chromatographic separation was performed on Rtx®-5MS capillary column (Restek, PA, USA), coated with 0.25 μm film of cross bond® 5 per cent diphenyl/ 95 per cent dimethyl polysiloxane; dimensions 30 m length × 0.25 mm internal diameter, 0.25 μm film thickness. Helium was used as the carrier gas at a flow rate of 44.5 cm/sec. The oven temperature programme was 80°C for 2 min, 80 to 250°C at 15°C/min and 10 min at 250°C. Two μl of sample was injected in 1:75 split ratio. The injector temperature and the interface temperature was 250°C. The mass spectrometer was operated in electron positive-mode ionization, with 70 eV ionization voltage. Ion source temperature was 250°C. The data were obtained in full scan mode (total ion current-TIC); mass range 35 - 500 amu. Identification of the compounds was done by comparing their mass spectra with the reference spectra in National Institute of Standards and Technology (NIST) library (http://chemdata.nist.gov/).
Molecular docking: Schrödinger Small-Molecule Drug Discovery Suite Release 2013-1 (Schrödinger 2013, New York, USA) and the products included therein were used for performing various molecular modelling studies. Glide version 5.9 was used to dock the compounds isolated from the active fraction, native ligand and known LasR inhibitors19 with two target proteins, LasI and LasR. A single structure file of AHL synthase, LasI from P. aeruginosa, is available in the Protein Data Bank (PDB; www.rcsb.org/) (PDB ID 1RO5). For LasR of P. aeruginosa, the structure showing LasR bound to its autoinducer (PDB ID 2UV0) was used. The protein structures were prepared using Protein Preparation wizard and subjected to Receptor Grid Generation using Glide 5.9. The generated grid was used for docking the compounds in the active site. Similarly, the structures of the compounds identified in fraction 5 were built in Maestro. All the structures were then subjected to LigPrep2.6, using the default settings. This set of molecules was subjected to docking in the active site of target proteins. For docking studies, Extra Precision (XP) mode was used. The docked poses were minimized and root mean square deviation (RMSD) to input molecular geometries were calculated.
High performance (HP) TLC fingerprinting: HPTLC was performed for fingerprinting of the crude extract (CEE) and the column fractions using two mobile phases; n-hexane:ethyl acetate::17:3 which is known to separate diterpenoids20 and a general separation system, toluene:ethylacetate:methanol:formic acid::8:2:1:1.HPTLC was performed on 10×10 cm aluminium plates coated with 0.25 mm layer of silica gel 60 F 254 (Merck, Germany); 20 μl of the extract (10 mg/ml in ethyl acetate) was applied as 8 mm wide and 8.0 mm apart, at a rate of 5 μl/sec, using a DESAGA Applicator AS 30 sample applicator (Sarstedt, Nümbrecht, Germany) equipped with a 100-μl Hamilton (USA) syringe. The plates were air dried, allowed to develop up to 80 mm in a pre-saturated (30 min) glass twin trough chamber containing 20 ml of the respective mobile phase. After development, plates were dried and visualized under a UV cabinet at 254 and 366 nm. The plates were scanned at 254 and 366 nm, using a HPTLC Densitometer CD 60, 230V, equipped with ProQuant software (Sarstedt, Nümbrecht, Germany). The plates were derivatized using Anisaldehyde sulphuric acid reagent21 and again scanned at 366 nm and 540 nm.
Statistical analysis: The experimental data were analyzed using paired t test, Wilcoxon signed rank test, one way analysis of variance (ANOVA) with the post hoc test, Dunnett's test, considering the level of significance (P<0.05). The statistical analysis software, SPSS 17.0 (Chicago, USA) was used.
Results
Concentration dependent anti-QS activity and IC50 value: Anti-QS activity increased with increasing concentration of CEE, from 1.0 to 10.0 mg/ml. Lower extract concentrations (≤ 0.5 mg/ml) did not show significant (P<0.05) inhibition. IC50 value for long acyl-HSL inhibition was calculated to be 4.48 mg/ml for A. baumannii, 4.79 mg/ml for P. aeruginosa.
Qualitative AHL detection, protease detection and typing of clinical isolates: In total, 36 isolates of P. aeruginosa and seven isolates of A. baumannii were obtained. Of all 36 isolates of P. aeruginosa, one isolate did not show AHL production, while all the isolates of A. baumannii were found to produce AHLs. Except one, all the clinical isolates showed protease activity. The clinical isolates of P. aeruginosa were classified into two groups, group I contained all the isolates except one, including the reference strain (PA01) and group II contained one isolate, labelled as C03. All group I isolates inhibited growth of the group II isolate; while C03 did not inhibit growth of group I isolates.
Effect of CEE on virulence of P. aeruginosa and biofilm formation of P. aeruginosa and A. baumannii: CEE was found to significantly (P<0.05) inhibit the production of the three virulence factors in the wild type as well as clinical isolates however, it was less than that of azithromycin (at 4 μg/ml concentration) (Fig. 1). P. aeruginosa showed increased biofilm density in the presence of CEE as compared to negative control (Table I). In case of A. baumannii, varied results were obtained for different clinical isolates (Table II). Azithromycin, the positive control was found to significantly (P<0.05) inhibit biofilm formation in P. aeruginosa, while it inhibited biofilm formation in A. baumannii, only at 72 h.
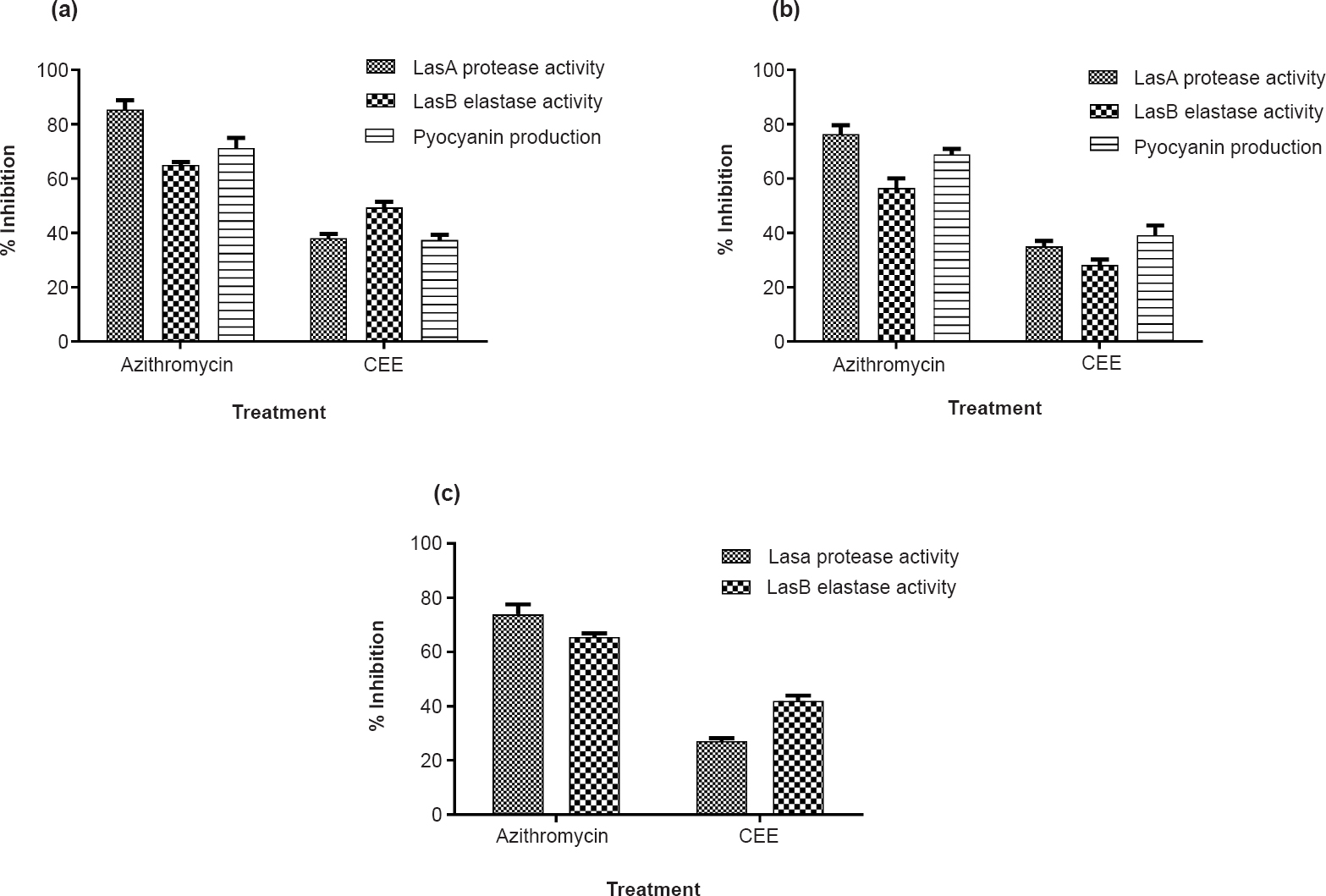
-
Virulence inhibition of P. aeruginosa: Per cent inhibition of the P. aeruginosa extracellular virulence factors (LasA protease, LasB elastase and pyocyanin) by CEE and azithromycin (positive control) in the test strains, PA01 (a), K04 (b), and C03 (c). Since C03 is a non-pigmenting strain, pyocyanin assay could not be performed. Mean ± SD values (n=9) are plotted.
Phytochemical analysis of CEE: Carbohydrates and steroids were detected, while amino acids, tannins, alkaloids, flavonoids, cardiac glycosides, anthroquinone glycosides and saponins were not detected.
Bioactivity of column fractions: Of all, fraction 5 (eluted with petroleum ether with 50% ethyl acetate) showed significant (P<0.05) anti-QS, anti-virulence and anti-biofilm activity at 1 mg/ml concentration (data not shown). As opposed to the crude extract (CEE), fraction 5 was also able to reduce biofilm formation in P. aeruginosa, suggesting that purification led to the separation of the compounds interfering with the anti-biofilm activity of the extract. It did not have any effect on bacterial growth at the above concentration, when tested using standard disc diffusion assay for antibacterial activity (data not shown).
GC-MS identification of compounds in fraction 5: Fig. 2 represents the compounds identified in fraction 5, along with their retention time (tR) on TIC chromatogram. tR1 and tR2 denote retention time for the compounds, when detected in non-derivatized and derivatized fraction, respectively. Before derivatization, three compounds with tR between 9 and 13 min were detected, while after derivatization, 10 compounds showing tR between 7 and 16 min were detected.
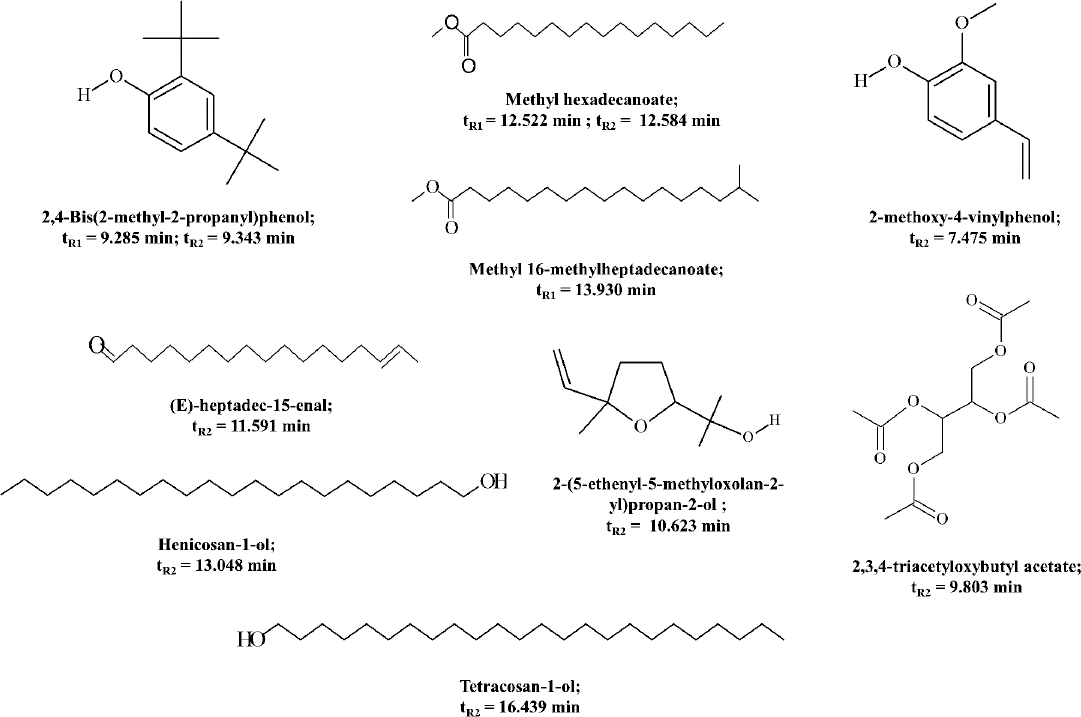
- Chemical structure, IUPAC name, and GC retention time (tR) of the compounds identified in fraction 5, obtained by comparing the mass spectra of analytes with the NIST mass spectral database. tR1 and tR2 represent the retention times for compounds detected in GC-MS analysis of non-derivatized and derivatized samples of fraction 5, respectively.
Docking scores: The summary of the docking results is given in Table III, showing the docking scores of compounds with the target proteins, LasI and LasR. Fig. 3 and 4 depict the docking poses of the native ligand, and the top ranking test compounds with LasI and LasR.
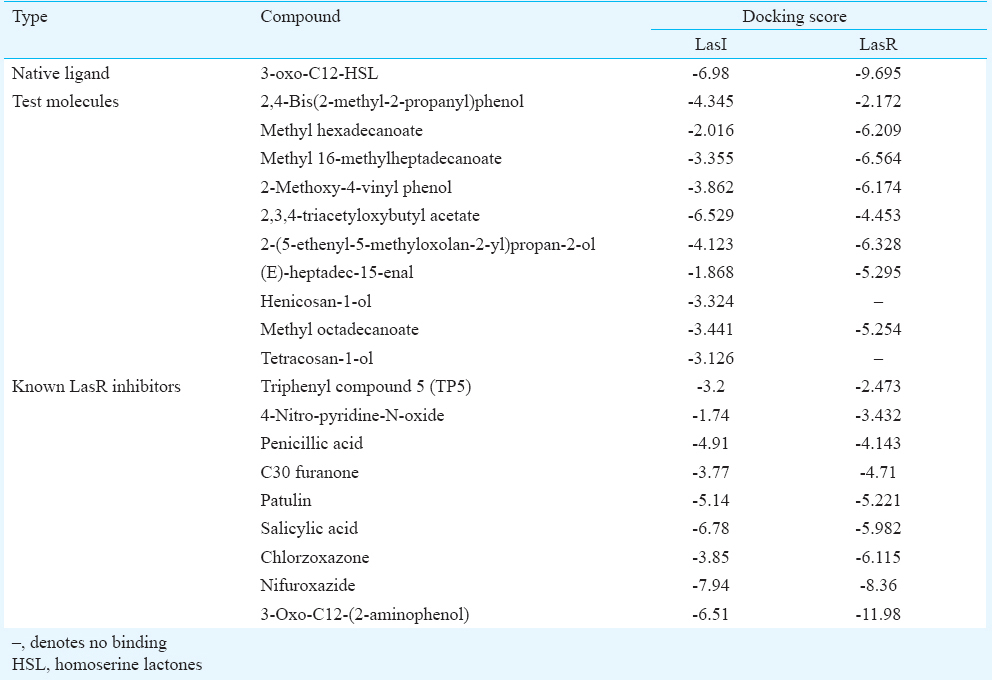
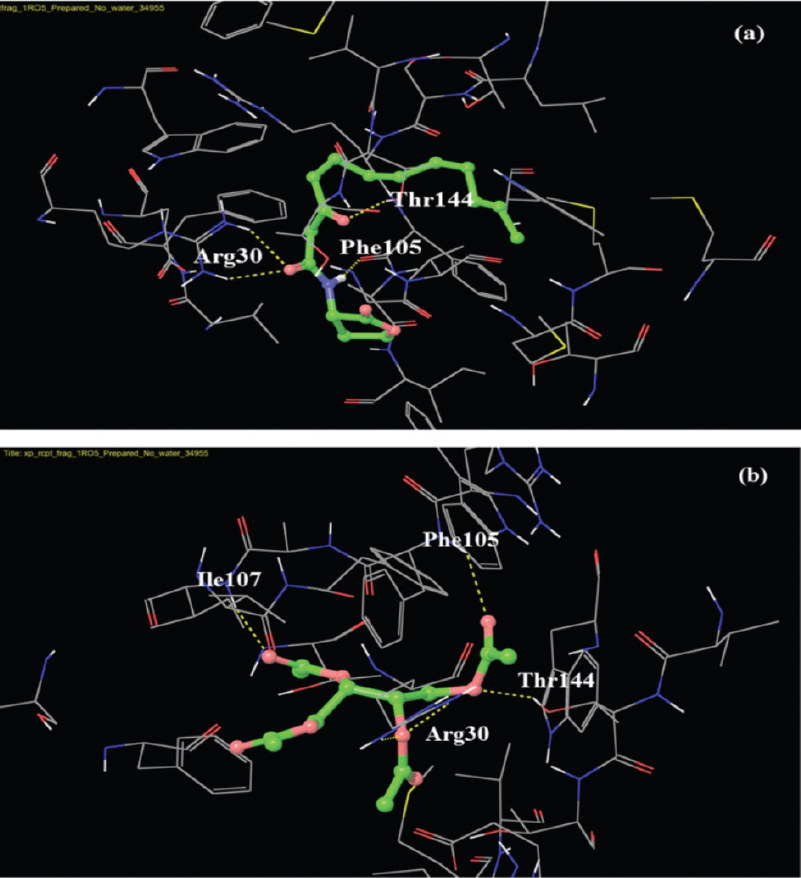
- Docking pose of 3-oxo-C12-HSL (homoserine lactores) (a) and 2,3,4-triacetyloxybutyl acetate (b) in the acyl-binding site of LasI. The ligands are shown as ball-and-stick model with hydrogen-bonds represented by yellow dotted lines; oxygen atoms in red and hydrogen atoms in white. The possibilities of hydrogen bond formation between the compound and amino acids of acyl site of LasI are presented.
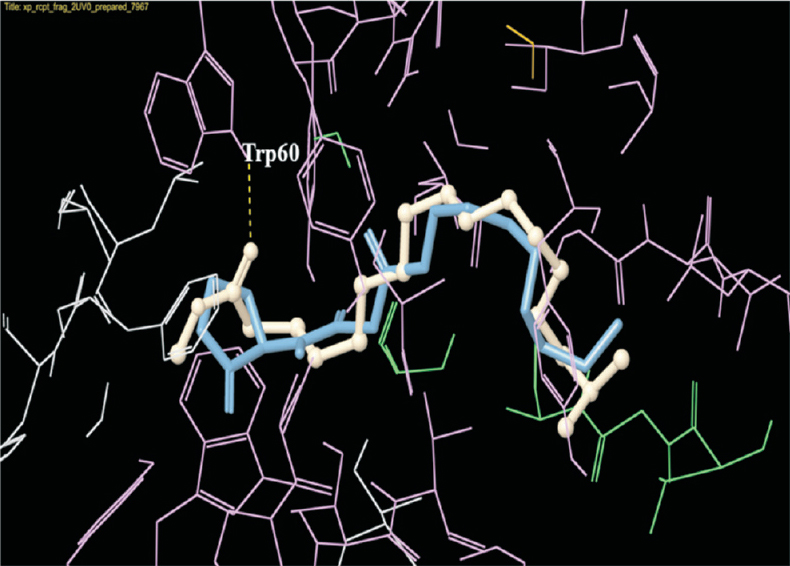
- Docking pose of 3-oxo-C12-HSL (blue) and methyl 16-methylheptadecanoate (white) in the active site of LasR. The ligands are shown as ball-and-stick model with H-bonds represented by the yellow dotted lines.
HPTLC fingerprinting: HPTLC separation of phytochemicals in CEE and column fractions in mobile phases I and II was visualized, and recorded. It showed the presence of terpenoids and sugars in accordance with the qualitative phytochemical data. The band pattern of column fractions showed clear separation of non-polar to polar compounds in fractions obtained by elution with non-polar 100 per cent petroleum ether to mid-polar 100 per cent ethyl acetate and upto polar 80 per cent methanol.
Discussion
In an initial screening study, the ethyl acetate extracts of the stem of T. cordifolia exhibited potential anti-QS activity7. In the present study, the ability of cold ethyl acetate extract of the plant was explored for attenuation of QS dependent virulence and biofilm development in the pathogens, P. aeruginosa and A. baumannii.
CEE at a concentration of 5 mg/ml was able to significantly inhibit the production of all three virulence factors (LasA protease, LasB elastase and pyocyanin) in the wild type and clinical isolates of P. aeruginosa. In a study by Adonizio22, among the plants tested, Conocarpus erectus and Bucida bucerus showed maximum inhibitory activity against QS in P. aeruginosa and were further tested for supression of virulence. Both plants showed significant decrease in LasA activity. C. erectus showed a significant effect on biofilm of P. aeruginosa PA01 strain. Such variations in the activity could be possible due to the chemical complexity of the plant extract, suggesting the need for further purification. In a similar study23, mangrove plants, belonging to Rhizophora spp., were found to significantly inhibit QS and QS regulated virulence and biofilm maturation in the wild type and drug resistant clinical isolates of P. aeruginosa. This study showed a potential source for QS inhibitors, which was able to inhibit virulence supporting the role of QS as an important target in anti-virulence therapy.
In P. aeruginosa, CEE significantly enhanced biofilm formation for the wild type strain as well as the clinical isolate K04, but there was no significant difference in the biofilm density of C03. According to Donlan24, with increase in surface roughness, there is an increase in microbial colonization, due to larger surface area and reduced shear forces; so the roughness provided by CEE may be relevant. Various sugars have been detected in the extracellular polymeric substance (EPS) of P. aeruginosa biofilm25, and glucose was found to promote biofilm formation26. Thus, the sugars present in CEE might have helped in larger production of EPS. Due to surface roughness and larger extracellular polymeric substance (EPS) production, thicker biofilms would have formed and the active compound/s might have been unable to cross the thick EPS matrix and exert its anti-QS effect. Additionally, the CEE is a mixture of numerous compounds which can have varying bioactivities; hence a role of some of its phytochemicals in enhancing biofilm formation may not be ruled out.
CEE showed significant decrease in biofilm density of the wild type strain of A. baumannii at various time intervals. For all the clinical isolates, except K06, K20 and K23, there was significant inhibition at 48 and 72 h. Variations in biofilm formation of the clinical strains of A. baumannii has been detected earlier27, however, the molecular basis of the differences or correlation with the biofilm formation and its associated cell properties could not be depicted. In the present study, variation in biofilm inhibition for wild type and clinical isolates was observed. Overall, CEE significantly inhibited biofilm in A. baumannii, suggesting a QS dependent mechanism of action.
As opposed to the crude extract (CEE), fraction 5 was also able to reduce biofilm formation in P. aeruginosa, suggesting that purification assisted in separation of the compounds interfering with the anti-biofilm activity of the extract. The active fraction 5 showed the presence of some non-polar, aliphatic compounds, such as fatty acids and their derivatives in GC-MS analysis. Unsaturated fatty acid such as cis-9-octadecenoic acid was reported to possess quorum quenching and anti-biofilm activity28. Also, branched chain fatty acids were found to affect biofilm formation of P. aeruginosa29. Hence, it was hypothesized that the fatty acids, hexadecanoic acid and octadecanoic acid, detected in the active fraction, might be responsible for anti-QS activity. However, when pure standards of these compounds were evaluated for QS inhibition, the fatty acids showed no inhibitory effect on the AHL production of the organisms under study (data not shown).
To predict the anti-QS compounds in fraction 5, the compounds were subjected to docking analysis with LasI and LasR proteins, and their docking scores along with bonding possibilities studied to predict the probable QS inhibitors. The top ranking compounds for LasR that showed docking scores around -6.0, included methyl 16-methylheptadecanoate, 2-(5-ethenyl-5-methyloxolan-2-yl) propan-2-ol, methyl hexadecanoate and 2-methoxy-4-vinylphenol. The native ligand of LasR, 3-oxo-C12-HSL was predicted to make hydrogen bonds with Trp-60, Asp-73, Tyr-56 and Ser-129 in LasR active site. Due to structural similarity of methyl 16-methylheptadecanoate with the acyl chain of 3-oxo-C12-HSL, their binding interactions were also similar to some extent. The compound, 2-(5-ethenyl-5-methyloxolan-2-yl) propan-2-ol showed the possibility of a covalent bond formation with the Cys-79, suggesting a probable covalent inhibitor of LasR.
In a study by Müh et al30 triphenyl compounds (TP1 to TP5) have been screened for their ability to inhibit QS using molecular docking with LasR followed by bioassay evaluation. Four compounds were found to act as agonists, while a single compound, TP5 exhibited antagonistic activity. The LasR-binding interactions of the above compounds have been studied, and it was predicted that the LasR agonists will make both Trp-60 and Asp-73 hydrogen bonds, while the antagonists will make only the Asp-73 hydrogen bond at the active site of LasR. In the present study, the known LasR inhibitor, 3-oxo-C12-(2-aminophenol) is predicted to form both Trp-60 and Asp-73 hydrogen bonds. Also, the top ranking test ligand, methyl 16-methylheptadecanoate is predicted to form Trp-60 hydrogen bond. In a study by Kim et al31, furanone derivatives that showed QS inhibitory activity, were found to make hydrogen bonds with Tyr-56, Leu-36 and Met-116. Hence, with further analysis on the binding interactions of different inhibitors, it will be possible to better understand the diverse mechanisms involved.
Most of the studies on QSIs have focussed on AHL analogues that act by competitive inhibition of LasR. However, LasI is also an important target protein for controlling quorum sensing, since it directly affects AHL production. Hence, in addition to LasR, the ligand molecules were also docked with LasI. Among the test molecules, 2,3,4-triacetyloxybutyl acetate showed the highest score. When the LasI-binding interactions of 2,3,4-triacetyloxybutyl acetate and 3-oxo-C12-HSL was compared, both compounds showed the possibilities of hydrogen bonds with Arg-30, Phe-105, Thr-144, and in addition to this, 2,3,4-triacetyloxybutyl acetate showed hydrogen bond formation with Ile-107. This indicated a strong possibility of the above test compound to be an antagonist of LasI synthase protein, which can result in decreased AHL production, and thus attenuate QS and its regulated phenotypes.
In conclusion, the study provides with some phytochemicals, isolated from the ethyl acetate extract of T. cordifolia stem, which can probably serve to interfere with the AHL-based quorum sensing in P. aeruginosa and A. baumannii. Future studies will focus on experiments to validate the anti-QS ability of these compounds, in their pure form.
Conflicts of Interest: None.
Acknowledgment
Authors thank Dr Barbara H. Iglewski, Department of Microbiology and Immunology, University of Rochester, New York, USA, for providing the strains, P. aeruginosa PA01 and E. coli MG4/pKDT17; Dr Robert J. C. McLean, Department of Biology, Texas State University, Texas, USA for providing the strain, C. violaceum ATCC12472. The authors acknowledge King Edward Memorial Hospital, Mumbai, India and Dr R. N. Cooper Hospital, Mumbai, India for providing the clinical isolates. The first author (VCG) acknowledges the Indian Council of Medical Research (ICMR), New Delhi, India, for providing Senior Research Fellowship.
References
- Targeting virulence: a new paradigm for antimicrobial therapy. Nat Chem Biol. 2007;3:541-8.
- [Google Scholar]
- Device associated nosocomial infections and patterns of antimicrobial resistance at a tertiary care hospital. J NTR Univ Health Sci. 2012;1:86-9.
- [Google Scholar]
- Prevalence of non-fermenting Gram negative bacilli and their in vitro susceptibility pattern in a tertiary care hospital of Uttarakhand: A study from foothills of Himalayas. Saudi J Heal Sci. 2013;2:108-12.
- [Google Scholar]
- Cell-to-cell signaling and Pseudomonas aeruginosa infections. Emerg Infect Dis. 1998;4:551-60.
- [Google Scholar]
- Isolation and characterization of an autoinducer synthase from Acinetobacter baumannii. J Bacteriol. 2008;190:3386-92.
- [Google Scholar]
- Evaluation of the potential of five medicinal plants to inhibit acyl homoserine lactone based quorum sensing in Pseudomonas aeruginosa and Acinetobacter baumannii. Int J Pharma Bio Sci. 2013;4:445-53.
- [Google Scholar]
- Azithromycin inhibits quorum sensing in Pseudomonas aeruginosa. Antimicrob Agents Chemother. 2001;45:1930-3.
- [Google Scholar]
- Assay of β-galactosidase. In: Experiments in molecular genetics. New York, USA: Cold Spring Harbor Laboratory; 1972. p. :352-5.
- Biofilms on indwelling urethral catheters produce quorum-sensing signal molecules in situ and in vitro. Appl Environ Microbiol. 1998;64:3486-90.
- [Google Scholar]
- Typing of Pseudomonas pyocyanea by pyocine production. J Pathol Bacteriol. 1966;91:339-45.
- [Google Scholar]
- Secreted LasA of Pseudomonas aeruginosa is a staphylolytic protease. J Biol Chem. 1993;268:7503-8.
- [Google Scholar]
- Roles of Pseudomonas aeruginosa las and rhl quorum-sensing systems in control of elastase and rhamnolipid biosynthesis genes. J Bacteriol. 1997;179:5756-67.
- [Google Scholar]
- Herbs, spices and medicinal plants used in hispanic traditional medicine can decrease quorum sensing dependent virulence in Pseudomonas aeruginosa. Int J Appl Res Nat Prod. 2008;1:9-15.
- [Google Scholar]
- Pseudomonas aeruginosa exotoxin pyocyanin causes cystic fibrosis airway pathogenesis. Am J Pathol. 2009;175:2473-88.
- [Google Scholar]
- Detection of biofilm formation among the clinical isolates of Staphylococci: an evaluation of three different screening methods. Indian J Med Microbiol. 2006;24:25-9.
- [Google Scholar]
- Preliminary phytochemical screening, Practical pharmacognosy techniques and experiments. (20th ed). New Delhi: Nirali Prakashan; 2010. p. :149-53.
- [Google Scholar]
- Determination of major phenolic acids, phenolic diterpenes and triterpenes in Rosemary (Rosmarinus officinalis L.) by gas chromatography and mass spectrometry. Acta Chim Slov. 2007;54:60-7.
- [Google Scholar]
- Computer-aided identification of recognized drugs as Pseudomonas aeruginosa quorum-sensing inhibitors. Antimicrob Agents Chemother. 2009;53:2432-43.
- [Google Scholar]
- Phytochemical methods. A guide to modern techniques of plant analysis (3rd ed). New Delhi: Springer; 1998. p. :292.
- Plant drug analysis. A thin layer chromatography atlas. Berlin Heidelerg, New York: Springer-Verlag; 1984.
- Anti-quorum sensing agents from South Florida medicinal plants and their attenuation of Pseudomonas aeruginosa pathogenicity [dissertation] Florida International University, USA; 2008.
- [Google Scholar]
- Antipathogenic potential of Rhizophora spp. against the quorum sensing mediated virulence factors production in drug resistant Pseudomonas aeruginosa. Phytomedicine. 2013;20:956-63.
- [Google Scholar]
- Alginate is not a significant component of the extracellular polysaccharide matrix of PA14 and PAO1 Pseudomonas aeruginosa biofilms. Proc Natl Acad Sci USA. 2003;100:7907-12.
- [Google Scholar]
- Quorum-sensing genes in Pseudomonas aeruginosa biofilms: their role and expression patterns. Appl Environ Microbiol. 2001;67:1865-73.
- [Google Scholar]
- Acinetobacter baumannii biofilms: variations among strains and correlations with other cell properties. J Microbiol. 2011;49:243-50.
- [Google Scholar]
- Cis-9-octadecenoic acid from the rhizospheric bacterium Stenotrophomonas maltophilia BJ01 shows quorum quenching and anti-biofilm activities. Biofouling. 2013;29:855-67.
- [Google Scholar]
- 12-Methyltetradecanoic acid, a branched-chain fatty acid, represses the extracellular production of surfactants required for swarming motility in Pseudomonas aeruginosa PAO1. Jpn J Infect Dis. 2012;65:126-31.
- [Google Scholar]
- A structurally unrelated mimic of a Pseudomonas aeruginosa acyl-homoserine lactone quorum-sensing signal. Proc Natl Acad Sci USA. 2006;103:16948-52.
- [Google Scholar]
- Furanone derivatives as quorum-sensing antagonists of Pseudomonas aeruginosa. Appl Microbiol Biotechnol. 2008;80:37-47.
- [Google Scholar]






