Translate this page into:
Allele-specific PCR for a cost-effective & time-efficient diagnostic screening of spinal muscular atrophy
Reprint requests: Dr Zilfalil Bin Alwi, Associate Professor, Department of Pediatrics, School of Medical Sciences, Universiti Sains Malaysia, 16150 Kubang Kerian, Kota Bharu, Kelantan, Malaysia e-mail: zilfalil@kb.usm.my
-
Received: ,
This is an open-access article distributed under the terms of the Creative Commons Attribution-Noncommercial-Share Alike 3.0 Unported, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
This article was originally published by Medknow Publications & Media Pvt Ltd and was migrated to Scientific Scholar after the change of Publisher.
Abstract
Background & objectives:
Genetic diagnosis of spinal muscular atrophy (SMA) is complicated by the presence of SMN2 gene as majority of SMA patients show absence or deletion of SMN1 gene. PCR may amplify both the genes non selectively in presence of high amount of DNA. We evaluated whether allele-specific PCR for diagnostic screening of SMA is reliable in the presence of high amount of genomic DNA, which is commonly used when performing diagnostic screening using restriction enzymes.
Methods:
A total of 126 blood DNA samples were tested in amounts ranging 80-200 ng, referred for the genetic diagnosis of SMA using both conventional PCR-RFLP and allele-specific PCR.
Results:
The results from both methods showed agreement. Further, allele-specific PCR was found to be a time-efficient and cost-effective method.
Interpretation & conclusions:
Our study demonstrated the accuracy of our allele-specific PCR and the results were comparable compatible with that of PCR-RFLP, indicating its practical application in SMA diagnostic screening.
Keywords
allele-specific PCR
PCR-RFLP
SMA diagnostic screening
SMN1
spinal muscular atrophy
Conventional PCR-RFLP for genetic diagnosis of spinal muscular atrophy (SMA)1 has been considered time consuming and expensive. It requires restriction enzyme (RE) digestion which uses a consi derably high amount of PCR product. If the amount of the PCR product is higher than necessary, this may lead to partial RE digestion resulting in the appearance of undigested PCR product on gel electrophoresis (i.e. false-negative results).
The responsible genes for SMA are Survival Motor Neuron (SMN) genes. The SMN genes consist of two highly identical genes; SMN1 (telomeric SMN) and SMN2 (centromeric SMN) which share over 99.8 per cent sequence homology over a 30 kb segment. SMN1 and SMN2 can be distinguished by only five nucleotides differences located in intron 6, exon 7, intron 7 and exon 82. Ninety five per cent of SMA patients showed an absence of SMN1 gene due to either deletion or conversion, thus demonstrating that SMN1, not SMN2, is the SMA-causing gene3. Thus, genetic diagnosis of SMA (i.e. detection of SMN1 deletion) was complicated by the presence of SMN2 because PCR may amplify the genes unselectively, especially in the presence of high amount of genomic DNA, while all patients carry SMN2 gene. Allele-specific PCR for the genetic diagnosis of SMA has been described elsewhere from as early as 19994–9. However, to the best of our knowledge studies involving high amount of DNA, the amount of which is routinely used for genetic diagnosis, provided conflicting results regarding chances of SMN2 mis-amplification. In addition, no study has been done to evaluate the cost-effectiveness and time-efficiency of this method over conventional PCR-RFLP. Using relatively larger sample size, we studied the reliability of allele-specific PCR by comparing the test results against conventional PCR-RFLP using high amount of genomic DNA.
Material & Methods
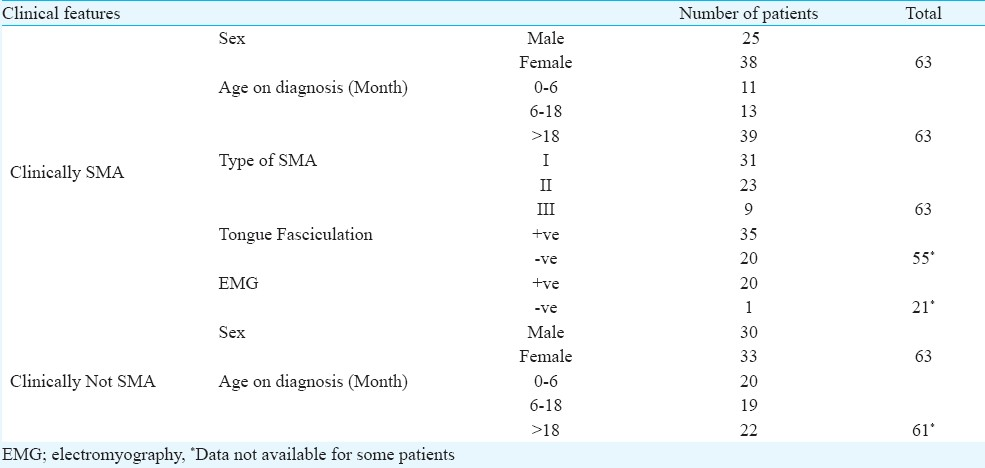
This study was carried out from 2003 to 2008 in the Department of Paediatric, School of Medical Sciences, Universiti Sains Malaysia, Kelantan. A total of 126 patients were randomly selected from the patients sent for SMA genetic diagnosis (Table I). Whole blood (3-5 ml) was collected from patients. Informed consent was taken prior to blood taking. The study protocol was approved by the Research Ethics Committee (Human) of the Universiti Sains Malaysia. Sample size was calculated using single proportion formula. Genomic DNA was extracted using commercially available kit (GeneAll Biotechnology Co. Ltd., Korea).
PCR-RFLP (Method A): All samples were analyzed twice, each using two different methods, method A and method B. Method A refers to the PCR-RFLP as previously described1. This method consisted of two steps, PCR amplification and enzyme digestion which used Dra1 restriction enzyme for exon 7 SMN.
Allele-specific PCR (Method B): Method B refers to allele-specific PCR using primer pairs described previously4, telSMNex7forw 5’-TTTATTTTCCTTACAGGGTTTC-3’ and telSMNint7rev 5’-GTGAAAGTATGTTTCTTCCACgTA-3’. Italic uppercase characters indicate the position of nucleotide difference between SMN1 and SMN2, while lowercase character indicates the position of a deliberate mismatch. The primers specifically amplify exon 7 of SMN1, not SMN2. This method consisted of only one PCR amplification step. Positive or negative interpretation was determined visually on agarose gel electrophoresis by the absence or presence of the SMN1 exon 7, respectively. To monitor the efficiency of PCR amplification, a housekeeping gene (β-globin) was used as a reference gene, with the primers 5-ACCTCACCCTGTGGAGCCAC-3 and 5-CTCACCACCAACTTCATCCAAG-3. One reaction of 20 μl of PCR mixture contained 80 - 200 ng of genomic DNA, 0.4 μl of 10 mM dNTPs, 1.2 μl of 25 mM MgCl2, 4.0 μl of 5x PCR buffer, 0.75U Taq DNA polymerase (Promega Corporation, Madison, USA) and 1.5 μl of each 10 pmol of an allele specific primer pairs and 0.5 μl of each 5 pmol of an internal control primer pairs (Sigma-Proligo, The Woodlands, TX, USA). The PCR cycles included an initial denaturation at 94°C for 7 min, followed by 33 cycles of 94°C for 1 min, 59.7°C for 1 min, 72°C 1 min before a final extension at 72°C for 7 min. The PCR product was then directly visualized under the UV light, using 2 per cent agarose gel. PCR product of SMN1 was visualized at the corresponding size of 307 bp, while that of β-globin was at the size of 240 bp.
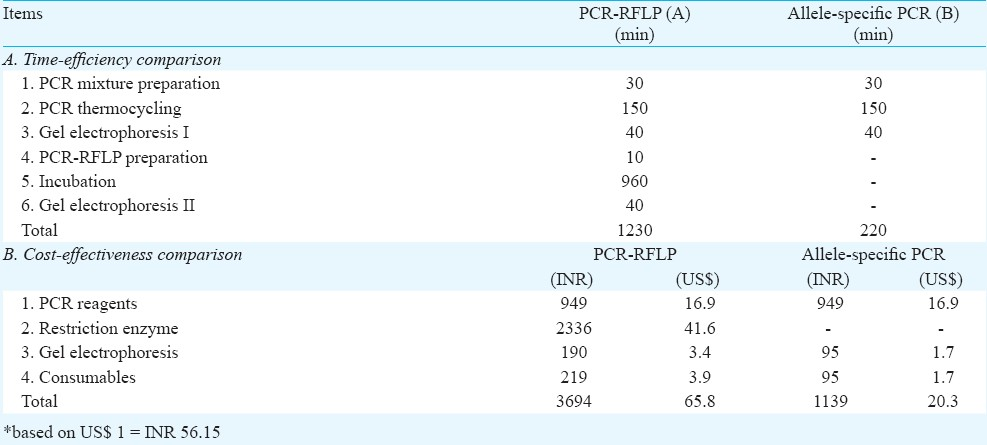
Cost and time evaluation: The cost and time for both methods were compared to evaluate the cost-effectiveness and time-efficiency of allele-specific PCR over PCR-RFLP.
Results & Discussion
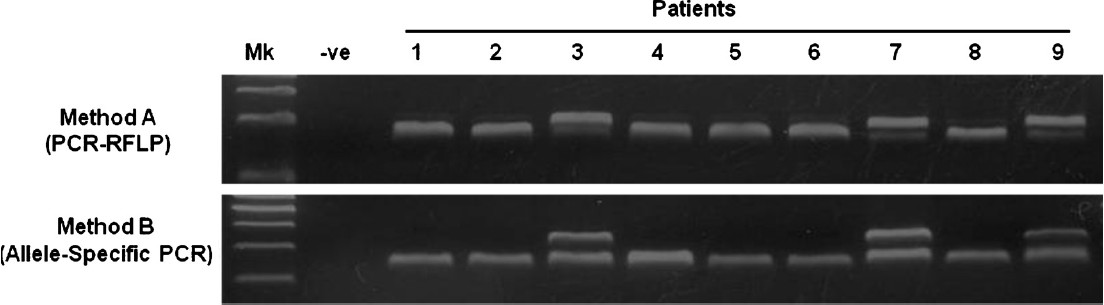
Of the 126 samples tested, 54 (43%) were found to have SMN1 exon 7 deletions. The findings (Fig.) from both methods were in complete agreement, suggesting that both methods performed with the same reliability. Evaluation of time-efficiency for both methods showed that method B was five times more rapid than method A. Evaluation on the cost-effectiveness showed that method B was more cost-effective (68%) compared to method A (Table II).
-
SMN1 (exon 7) deletion analysis using method (A) and (B). In method (A), deletion or non-deletion is indicated by the absence or presence of the first band (188 bp). The second band (164 bp) shows the presence of SMN2 (exon 7). In method (B), deletion or non-deletion is indicated by the absence or presence of the first band (307 bp). The second band (240 bp) indicated the presence of reference gene (β-globin). Molecular markers were electrophoresed in the “Mk” lane and a control PCR product in the “–ve” lane. Patients 1, 2, 4, 5, 6, 8 show deletion of SMN1. Patients 3, 7, 9 show non-deletion of SMN1.
We have been using the primers described by Feldkötter et al4 for gene-specific copy number analysis of SMN1 and SMN2 as a diagnostic procedure in our laboratory and found that our results were consistent1011. However, the experiments were done with relatively low amount of DNA, by which the chance for SMN2 mis-amplification was very small.
In this study, 54 samples showed deletion in SMN1 gene exon 7. Among the remaining 72 patients without SMN1 deletion, nine were categorized as clinically SMA (Table I). Patients without apparent deletion of SMN1 may not be excluded from the SMA diagnosis since possibility remains that they might carry point mutation which can only be identified through DNA sequencing.
Our analyses showed that allele-specific PCR was a time-efficient and cost-effective method. It may also reduce the risk for experimental errors since it involves fewer steps.
The main differences of the method described here compared to other similar methods is the design of the primers, the usage of conventional PCR method and the usage of a relatively higher amount of genomic DNA than the amount which is routinely used for SMA genetic diagnosis. Using primer pairs described by Feldkotter et al4. We could specifically amplify SMN1 in the existence of relatively high DNA amount (80 - 200ng), without mis-amplifying SMN2, thus enabling the use of the method for routine genetic testing of SMA. However, the use of higher amount of DNA in this study has not provided evidence if the test is still reliable in the presence of much lower DNA amount as described elsewhere58. This could be a major hurdle for applications such as preimplantation genetic diagnosis (PGD).
The primers were selected because these fulfilled the criteria for highly-efficient allele-specific amplification for utilizing two nucleotides differences between SMN1 and SMN2 in exon 7 (c.840C>T) and intron 7 (c.888+214A>G) and a deliberate primer mismatch simultaneously in one PCR reaction. SMN1 carries the C and A in its exon 7 and intron 7, respectively. The forward primer (telSMNex7forw) incorporated the C at its first nucleotide at the 3’ end, while the reverse (telSMNint7rev) combined an incorporation of A at its 2nd nucleotide before the 3’ end and a deliberate mismatch at its 3rd nucleotide before the 3’end (G instead of A). Newton et al12 showed that a deliberate mismatch near to the primer's 3’end increased its amplification specificity. Therefore, the primer pair described by Feldkotter et al4 contained three characteristics, a specific forward, a specific reverse and a deliberate mismatch.
Similarly, specific primers may be designed to detect the presence or absence of SMN1 exon 8. However, we concentrated only on exon 7 of the SMN1 gene in this study because it is the only region with the clinical significance for the diagnostic screening of SMA13. The summary of the previous studies were shown in Table III.

In conclusion, our study demonstrated the reliability of allele-specific PCR for diagnostic screening of SMA. The accuracy of this method was comparable with that of PCR-RFLP, and it was cost-effective. Thus, it can be applied to routine diagnostic screening of SMA.
This study was funded by the Universiti Sains Malaysia Short Term Grant (Grant No.304/PPSP/6131463). Authors thank Dr Alida Harahap of the Eijkman Institute, Jakarta, Indonesia for the provision of β-globin primers.
References
- PCR-based DNA test to confirm clinical diagnosis of autosomal recessive spinal muscular atrophy. Lancet. 1995;345:985-6.
- [Google Scholar]
- Structure and organization of the human survival motor neurone (SMN) gene. Genomics. 1996;32:479-82.
- [Google Scholar]
- Identification and characterization of a spinal muscular atrophy-determining gene. Cell. 1995;80:155-65.
- [Google Scholar]
- Quantitative analyses of SMN1 and SMN2 based on realtime light-cycler PCR: fast and highly reliable carrier testing and prediction of severity of spinal muscular atrophy. Am J Hum Genet. 2002;70:358-68.
- [Google Scholar]
- Allele-specific amplification for pre-implantation genetic diagnosis (PGD) of spinal muscular atrophy. Prenat Diagn. 2001;21:498-503.
- [Google Scholar]
- Allele-specific amplification for the diagnosis of autosomal recessive spinal muscular atrophy. Clin Chem Lab Med. 1999;37:133-5.
- [Google Scholar]
- Allele-specific amplification of exon 7 in the survival motor neuron (SMN) genes for molecular diagnosis of spinal muscular atrophy. Genet Test. 2003;7:325-7.
- [Google Scholar]
- Comparison of PCR-RFLP with allele-specific PCR in genetic testing for spinal muscular atrophy. Genet Test. 2003;7:277-81.
- [Google Scholar]
- Evaluation of an in-house protocol for prenatal molecular diagnosis of SMA in Chinese. Clin Chim Acta. 2008;398:78-81.
- [Google Scholar]
- SMN2 and NAIP gene dosages in Vietnamese patients with spinal muscular atrophy. Pediatr Int. 2008;50:346-51.
- [Google Scholar]
- Combination of SMN2 copy number and NAIP deletion predicts severity in spinal muscular atrophy. Brain Dev. 2009;31:42-5.
- [Google Scholar]
- Analysis of any point mutation in DNA. The amplification refractory mutation system (ARMS) Nucleic Acid Res. 1989;17:2503-16.
- [Google Scholar]
- SMN1 exon 7 deletion analysis may be necessary and sufficient for the diagnosis of spinal muscular atrophy. J Neurogenet. 2011;25:15-6.
- [Google Scholar]






