Translate this page into:
AdeR-AdeS mutations & overexpression of the AdeABC efflux system in ciprofloxacin-resistant Acinetobacter baumannii clinical isolates
For correspondence: Dr. Abdollah Ardebili, Department of Microbiology, Golestan University of Medical Sciences, Gorgan, Iran e-mail: ardebili_abdollah57@yahoo.com
-
Received: ,
This is an open access journal, and articles are distributed under the terms of the Creative Commons Attribution-NonCommercial-ShareAlike 4.0 License, which allows others to remix, tweak, and build upon the work non-commercially, as long as appropriate credit is given and the new creations are licensed under the identical terms.
This article was originally published by Medknow Publications & Media Pvt Ltd and was migrated to Scientific Scholar after the change of Publisher.
Abstract
Background & objectives:
Overexpression of efflux pumps is a cause of acquired resistance to fluoroquinolones in Acinetobacter baumannii. The present study was done to investigate the presence and overexpression of AdeABC efflux system and to analyze the sequences of AdeR-AdeS regulatory system in ciprofloxacin-resistant A. baumannii isolates.
Methods:
Susceptibility of 50 clinical A. baumannii isolates to ciprofloxacin, imipenem, ceftazidime, cefepime and gentamicin antimicrobials was evaluated by agar dilution method. Isolates were screened for the evidence of active efflux pump. Isolates were also examined for adeR-adeS and adeB efflux genes by polymerase chain reaction (PCR). The adeR and adeS regulatory genes were sequenced to detect amino acid substitutions. Expression of adeB was evaluated by quantitative reverse-transcriptase PCR.
Results:
There were high rates of resistance to ciprofloxacin (88%), ceftazidime (88%), cefepime (74%) and imipenem (72%) and less resistance rate to gentamicin (64%). Phenotypic assay showed involvement of active efflux in decreased susceptibility to ciprofloxacin among 16 isolates. The 12.27-fold increase and 4.25-fold increase were found in adeB expression in ciprofloxacin-full-resistant and ciprofloxacin-intermediate-resistant isolates, respectively. Several effective mutations, including A91V, A136V, L192R, A94V, G103D and G186V, were detected in some domains of AdeR-AdeS regulators in the overexpressed ciprofloxacin-resistant isolates.
Interpretation & conclusions:
The results of this study indicated that overexpression of the AdeABC efflux pump was important to reduce susceptibility to ciprofloxacin and cefepime in A. baumannii that, in turn, could be triggered by alterations in the AdeR-AdeS two-component system. However, gene expression alone does not seem adequate to explain multidrug resistance phenomenon. These results could help plan improved active efflux pump inhibitors.
Keywords
Acinetobacter baumannii
adeB
adeR
adeS
ciprofloxacin resistance
One of the Gram-negative pathogens that has acquired epidemiological importance among nosocomial infections is Acinetobacter baumannii1. This pathogen affected mainly patients with impaired host defenses in the Intensive Care Unit and burn wards. It has been implicated in a wide range of infections, including bacteraemia, wound infection, ventilator-associated pneumonia (VAP) and meningitis2. Particularly, the emergence and distribution of A. baumannii isolates with multiple drug-resistance (MDR-AB) or extensively drug-resistance in Iran34 and other parts of the world5 have become of great concern since we have now rather few treatment options against A. baumannii infections. Developed in the 1980s, fluoroquinolones showed potent activity against Acinetobacter strains and had even a better effect than the extended-spectrum cephalosporins or aminoglycosides6. However, resistance to these antibiotics rapidly arose among in A. baumannii clinical isolates as a result of extensive or unnecessary use in different medical settings worldwide789.
The best-known mechanism of resistance to quinolones in A. baumannii is spontaneous mutations in the quinolone resistance-determining region (QRDR) of gyrA and parC genes, encoding DNA gyrase and topoisomerase IV, respectively89. On the other hand, overexpression of efflux pumps is also a source of acquired resistance to fluoroquinolones in A. baumannii. AdeABC is the first and the major discovered efflux system in A. baumannii and belongs to the resistance-nodulation-cell division (RND) superfamily transporter10. The adeABC operon encodes the AdeA membrane fusion protein, the multidrug transporter protein AdeB and the AdeC outer membrane protein. Expression of adeABC is closely regulated by the AdeR-AdeS two-component system (TCS)11. Constitutive overexpression of the adeABC efflux system has been shown to be caused either by the ISaba-1 insertion upstream of the adeABC operon or by single- or multiple-point mutations in adeR and adeS genes. In these circumstances, due to decreased intracellular antibiotic concentration, A. baumannii becomes resistant to not only fluoroquinolones but also aminoglycosides, tetracyclines, chloramphenicol and β-lactams1112.
Single nucleotide polymorphisms (SNPs) are the most abundant form of genetic variations in closely related microbial strains or isolates. DNA sequencing studies detecting polymorphisms and comparing distinct drug-susceptible and drug-resistant strains improve our understanding of the evolutionary mechanisms of bacteria at the genomic levels and facilitate the development of next-generation antimicrobial agents. In this study, analysis of adeB gene expression was performed to evaluate the correlation between the active AdeABC efflux system and ciprofloxacin resistance in A. baumannii clinical isolates. In addition, sequencing analysis of the AdeR-AdeS TCS was performed to explore the role of mutations in the regulators to overexpression of AdeABC.
Material & Methods
Bacterial isolates: A. baumannii clinical isolates included in this study were obtained from the Motahari Burn and Reconstruction Center, a subset of Iran University of Medical Sciences (IUMS), in Tehran during 2012-2013. Bacterial isolates were initially identified by conventional biochemical methods in the department of Microbiology, School of Medicine, IUMS, Tehran, Iran, and then, species identification was performed by the polymerase chain reaction (PCR) amplification of the intrinsic blaOXA-51-like carbapenemase gene and sequencing13.
In vitro susceptibility testing: Minimum inhibitory concentration (MIC) value of five antibiotics (Mast, Merseyside, UK), including ciprofloxacin (0.015-128 μg/ml), imipenem (0.06-128 μg/ml), ceftazidime (0.25-128 μg/ml), cefepime (0.25-128 μg/ml) and gentamicin (0.06-128 μg/ml) was determined by the agar dilution method based on the recommendations of the Clinical and Laboratory Standards Institute14. Pseudomonas aeruginosa ATCC 27853 and Escherichia coli ATCC 25922 strains were used as controls. For determination of the presence of efflux system, the effect of the efflux inhibitor carbonyl cyanide 3-chlorophenylhydrazone (CCCP) (Sigma-Aldrich, United Kingdom) (final concentration 25 μg/ml) on ciprofloxacin MIC of isolates was tested as described previously15.
Conventional polymerase chain reaction (PCR) and sequence analysis: Analysis of the adeR-adeS locus was performed by PCR and sequencing. The primers specific for the genes encoding the transporter protein adeB and TCS comprising adeS and adeR are listed in Table I. DNA from the prepared isolates of A. baumannii was extracted using Genomic DNA Purification Kit (Fermentas; Vilnius, Lithuania). Amplification reactions of three genes were performed with the parameters described previously1617. The PCR products were analyzed by an ABI 3730XL DNA Analyzer (Applied Biosystems Inc., USA). The obtained sequences results were examined by using the NCBI BLAST program (http://www.ncbi.nlm.nih.gov/BLAST/). Multiple-sequence alignment of the deduced peptide sequences was performed using the ClustalW2 software program at the European Bioinformatics Institute website (http://www.ebi.ac.uk).
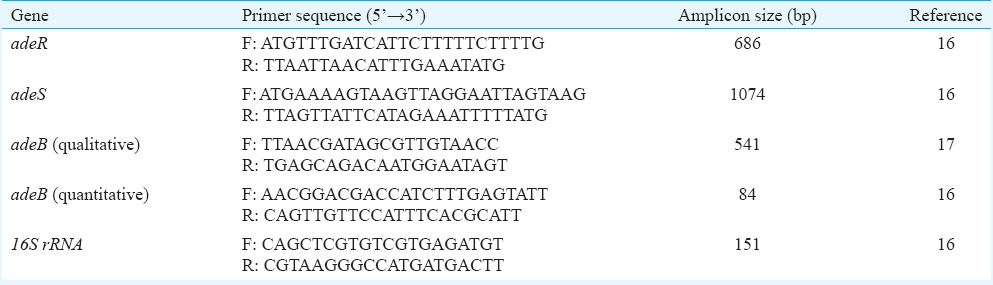
Quantitative real-time polymerase chain reaction (PCR) of adeB gene: Efflux-positive isolates were assessed for the expression of adeB gene. 16S rRNA was used as a housekeeping gene to normalize levels of adeB transcripts. Oligonucleotide primer sequences used for adeB and 16S rRNA are shown in Table I. Total RNA was initially extracted (Total RNA Purification Kit; Jena Bioscience, Germany) from cultures grown in the mid-log phase of growth in Luria-Bertani broth (Merck, Darmstadt, Germany) and then contaminating DNA was removed by RNase-free DNase I (Fermentas, Thermo Fisher Scientific Inc., Vilnius, Lithuania). The concentrations of RNA in each sample were quantified with a spectrophotometer at 260 and 280 nm. DNAse-treated RNA (2.5 μg) was reverse transcribed into cDNA using the CycleScript RT PreMix Kit (Bioneer, Korea) and 80 pM random hexamer (dN6) (Bioneer, Korea). Reverse-transcriptase PCR (RT-PCR) was performed by using a 2× GreenStar Master Mix Kit (Bioneer, Korea) on a Corbett Rotor-Gene 6000 real-time rotary analyzer (Corbett Life Science, Australia). A typical RT-PCR sample (25 μl) contained 11 μl of PCR Master Mix, 1 μl of cDNA, 0.5 μl of 0.8 μM solutions of both forward and reverse gene-specific primers and 12 μl of double distilled water. Real-time run protocol was the same as previously described16. Each sample was run in triplicate. A critical threshold cycle (CT) value was used to represent adeB transcripts quantitatively. The △CT for adeB transcripts was calculated against that for the 16S rRNA gene, and the △△CT was calculated against that for the ciprofloxacin-susceptible strain, A. baumannii ATCC 19606. The adeB relative expression was calculated by the 2−△△CT method.
Statistical analysis: SPSS 18 for Windows (SPSS Inc., Chicago, IL, USA) was used for statistical analysis. Differences in the mean expression level of adeB transcripts in the clinical isolate groups against that the A. baumannii ATCC 19606 control strain were assessed by independent samples t test. Categorical variables, including antibiotic resistance pattern, mutation in the adeR-adeS locus and adeB expression, were compared using the Chi-square test. The relationship between ciprofloxacin MIC values with the number of adeR and adeS mutations and with the expression level of adeB gene was assessed by calculating Spearman's correlation coefficient.
Results
Bacterial isolates, susceptibility profiles and effects of carbonyl cyanide 3-chlorophenylhydrazone (CCCP): During a nine-month period, a total of 50 single patient isolates of A. baumannii were recovered from hospitalized burn patients. Eighteen per cent (9 out of 50) and 82 per cent (41 out of 50) of the bacterial isolates were originated from blood and wound specimen cultures, respectively.
High rates of resistance to ciprofloxacin (88%), ceftazidime (88%), cefepime (74%) and imipenem (72%) were observed. However, isolates showed less resistance to gentamicin (64% resistant). Forty isolates (80%) were classified as MDR-AB based on non-susceptibility to >1 agent in ≥3 antimicrobial categories18. The MIC determinations of all test antimicrobials against A. baumannii isolates are shown in Table II. The MIC50 and the MIC90 values for ceftazidime were both <128 μg/ml, higher than those of other antibiotics tested, while imipenem represents the lowest determinations of MIC50 (32 μg/ml) and MIC90 (64 μg/ml).
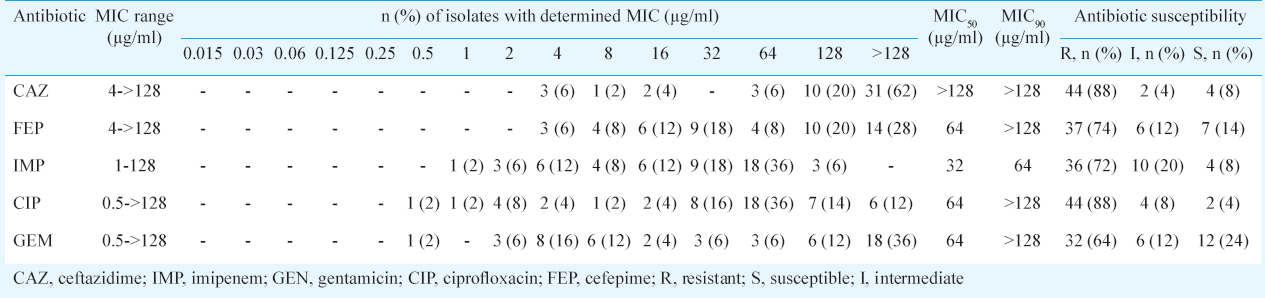
In the phenotypic assay for the detection of efflux phenotype, CCCP reduced MIC of ciprofloxacin from a 4- to >16-fold in 32 per cent (16 out of 50) of A. baumannii isolates, suggesting a putative efflux mechanism. All efflux-positive A. baumannii isolates in this study were ciprofloxacin-intermediate (MIC=2 μg/ml) and ciprofloxacin-full resistant (MIC=4 to ≥128 μg/ml) (Table II).
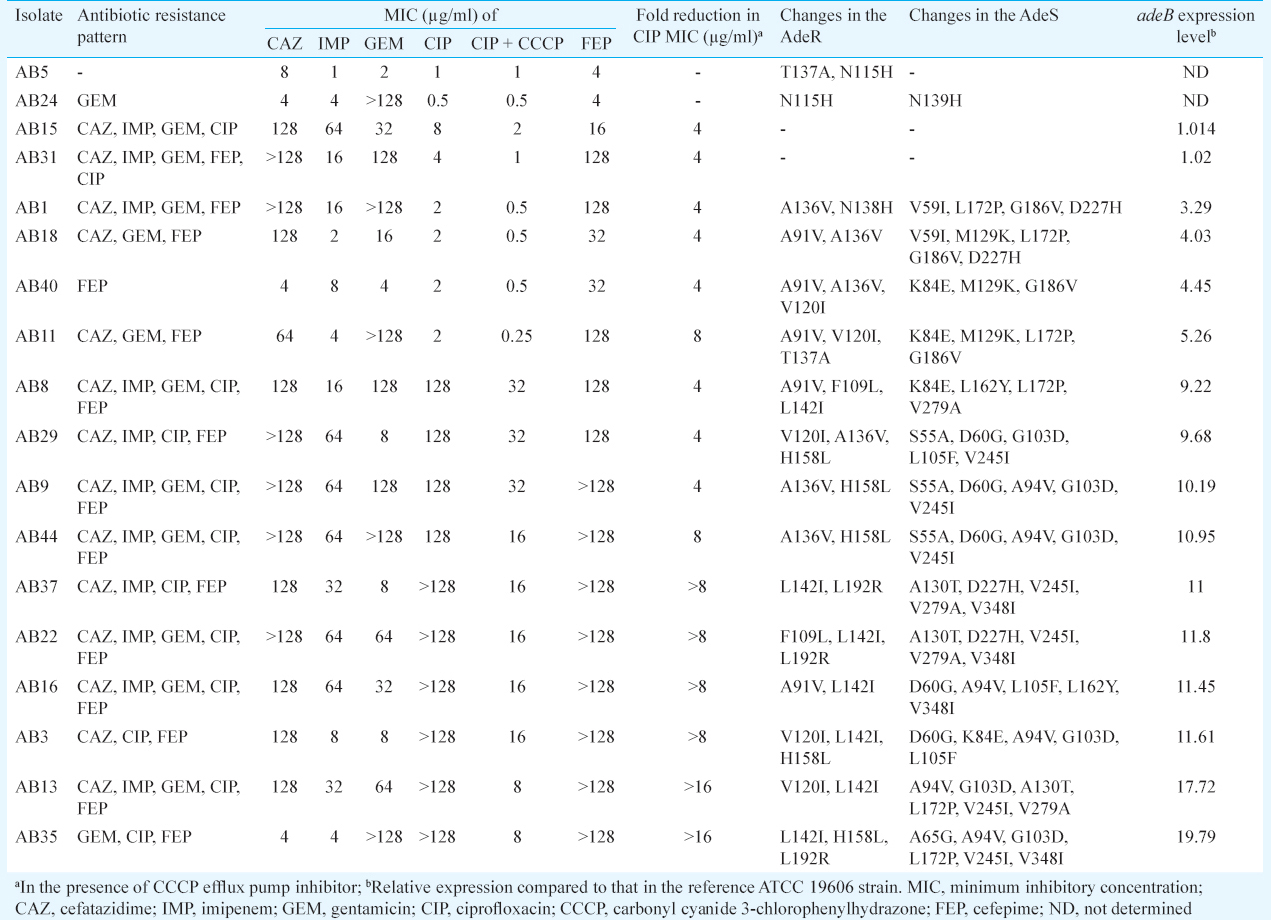
Sequencing of the adeR-adeS regulatory system: All 18 clinical A. baumannii isolates, including 16 efflux-positive and two ciprofloxacin-susceptible isolates, were found to carry chromosomal adeB, adeR and adeS genes, simultaneously. The sequencing analysis of the PCR products showed that multiple-point mutations were common in adeR-adeS locus. Both ciprofloxacin-susceptible isolates were found to have two mutations each. AB5 had mutations located at the T137A and N115H positions in the AdeR. Isolate AB24 had mutations located in the N115H and N139H positions within the AdeR and AdeS, respectively (Table III). Multiple-sequence alignment of AdeR and AdeS from clinical isolates in comparison to A. baumannii ATCC 17978 is shown in Fig. 1. None of the isolates possessed the P116L and T153M substitutions in the AdeR and AdeS, respectively, which were identified to cause altered expression of adeB11. However, some mutations associated with the adeABC overexpression and also SNPs were observed in 87.5 per cent (14 out of 16) of efflux-positive isolates (Table III).
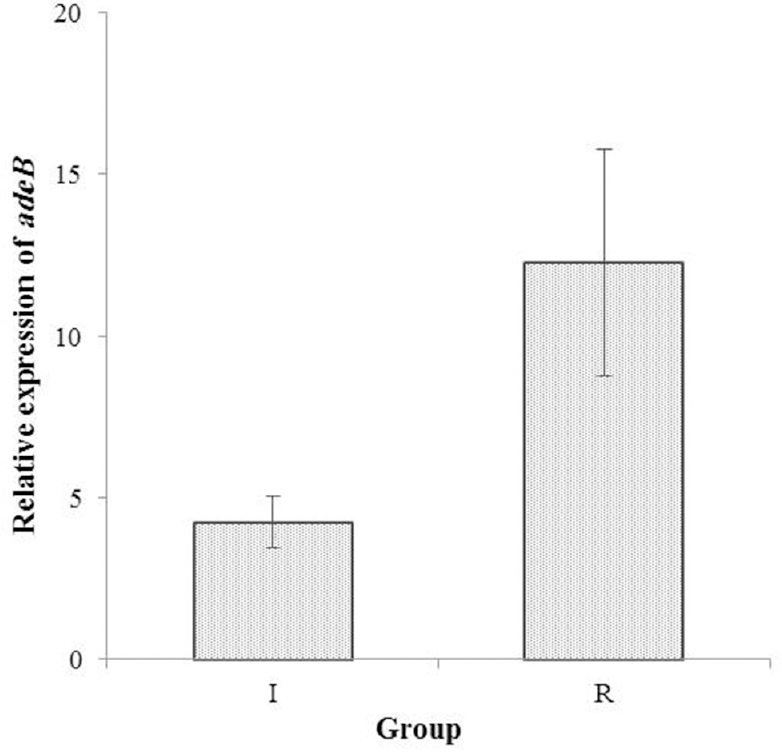
- Expression of adeB mRNA transcripts as evaluated by real-time polymerase chain reaction. adeB expression in two ciprofloxacin-full resistant (R) and ciprofloxacin-intermediate (I) groups of Acinetobacter baumannii was compared relative to that in the reference ATCC 19606 strain. The error bars represent the standard deviation for the average of results from three independent experiments.
Gene expression analysis of adeB: The 3.29-fold to 19.79- fold increases in the adeB expression level were detected in 14 efflux-positive isolates in comparison to ciprofloxacin-susceptible A. baumannii control strain. Fig. 2 shows the relative mean±standard deviation expression of adeB mRNA transcript as assessed by RT-PCR. Comparing the relative quantification of adeB expression in the clinical isolates to those in the ATCC 19606 control strain, 12.27±3.5 and 4.25±0.82 times more adeB transcripts were observed in ciprofloxacin-full-resistant and ciprofloxacin-intermediate-resistant A. baumannii, respectively (P <0.001). All of the 14 isolates with higher levels of adeB expression than the reference strain exhibited alterations in adeR and adeS genes. In terms of drug resistance pattern, 12 of the 16 efflux-positive isolates (75%) showed multidrug resistance, of which two isolates had no adeB overexpression.
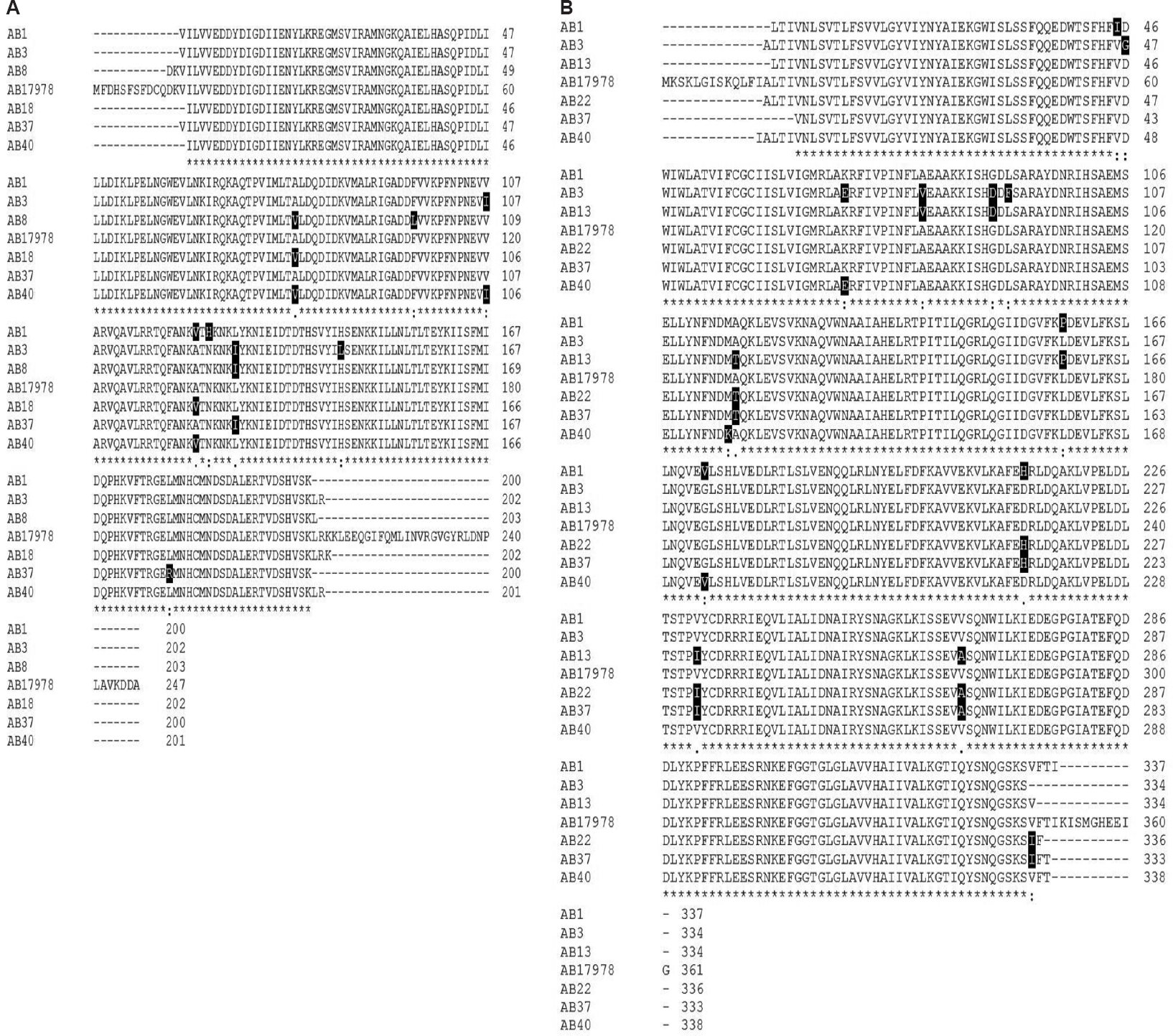
- Multiple-sequence alignment of AdeR (A) and AdeS (B) from Acinetobacter baumannii ATCC 17978 and clinical isolates. Sequence alignment was generated using ClustalW2 software program. The deduced amino acid sequence is designated in a single letter code. Asterisks indicate identical residues, colons indicate strongly similar residues and dots indicate weakly similar residues. Amino acid substitutions in AdeS and AdeR were highlighted in black. Some mutations leading to constitutive expression of AdeABC pump in clinical isolates such as A91V, A136V and L192R in AdeR and A94V, G103D and G186V in AdeS are indicated.
Nucleotide sequence accession number: A selection of the adeR and adeS sequences reported in this study has been submitted to NCBI and deposited in the GenBank data library under the following accession numbers: KY000415, KY000416, KY000417, KY000418, KY000419, KY000420, KY000421, KY000422, KY000423, KY000424, KY000425, KY000426 and KY000427.
Discussion
Antimicrobial resistance is the most important challenge in treating infections due to A. baumannii2. The necessity for effective therapy regimens to control MDR-AB outbreaks in hospital has been well emphasized45. An alarming trend of increase in MDR-AB resistance towards different classes of antibiotics has been shown in Tehran, Iran. In particular, Bahador et al19 revealed an increase in MIC values to all test antimicrobials among MDR-AB isolates, between 2006 and 2011. Over a period of five years, MIC50 for the majority of and MIC90 for all antimicrobials tested also rose. Similar to this, in our study also MIC range of all test antibiotics against the majority of isolates was high. Other studies also have shown a significant increase in most antimicrobial agents in the world202122. Generally, our findings were consistent with these studies that showed A. baumannii exhibited high-level resistance to all the first-line antibiotics, including anti-pseudomonal cephalosporins (ceftazidime or cefepime), anti-pseudomonal carbapenems (imipenem or meropenem), fluoroquinolones (ciprofloxacin or levofloxacin) and aminoglycosides (gentamicin or amikacin). In such circumstances, it is important to determine antibiotic susceptibility profiles and exert severe infection control measures to overcome nosocomial MDR-AB strains.
The importance of the AdeABC efflux pump in conferring MDR-AB has been discussed1023. In a small-scale study, Higgins et al12 found a 20-fold increase in the adeB transcripts of two high-level ciprofloxacin-resistant (both MIC ≥256 μg/ml) and ofloxacin-resistant (both MIC ≥64 μg/ml) isolates. MICs of non-fluoroquinolone antibiotics, including meropenem, were also found to have risen in the both adeB-overexpressed strains. In contrast, Bratu et al24 suggested that increased level of adeB expression by itself did not have a major role in fluoroquinolone resistance, but other mechanisms must be accounted. Consistent with Higgins et al12, we found a significant correlation between ciprofloxacin resistance and upregulated adeB. Fourteen of the 16 efflux-positive isolates overexpressed adeB, while 10 and four of 14 hyper-expressing isolates showed full resistance and intermediate resistance to ciprofloxacin, respectively. In addition, it was found that isolates with full resistance to ciprofloxacin had significantly higher adeB expression level than intermediate-resistant isolates. Taken together, the results suggested that upregulation of the adeB gene might be important as much as mutational mechanism in gyrA and/or parC genes to decreased susceptibility to fluoroquinolones in A. baumannii.
It seems that reduced susceptibility to cefepime should be mediated partly by the AdeABC efflux pump. Bratu et al24 found that in the absence of cephalosporinase activity, AdeABC efflux system was responsible for the reduced susceptibility to cefepime, and in the presence of an effective cephalosporinase, efflux-based mechanisms played a secondary role. Similar to what was observed for ciprofloxacin, our results indicated a significant association between cefepime resistance and AdeABC expression; increased level of the adeB expression was present along with higher resistance level to cefepime (MIC ≥128 μg/ml) in a significant percentage of efflux-positive isolates. However, similar to previous studies1225, resistance to three remaining antibiotics, imipenem, ceftazidime and gentamicin, was not significantly associated with the overexpression of adeB gene, indicating involvement of mechanisms other than the adeB expression to develop multidrug resistance.
AdeR-AdeS is an example of TCS that regulates strongly the expression of adeABC efflux pump in response to stimuli1026. In the present study, mutations in the adeR-adeS operon were investigated to identify the amino acid substitutions affecting AdeABC expression. No changes were found in adeR and adeS genes of AB15 and AB31; the isolates had adeB expression levels equal to the susceptible reference strain. Considering to the restoration of ciprofloxacin activity with the addition of CCCP, there must be other efflux systems but AdeABC, including AdeIJK, AdeFGH, AbeM and AbeS, to remove fluoroquinolones from the cell27.
In AdeR, three SNPs leading to increased expression of AdeABC have been reported: D20N located in the D box of the phosphorylation site28, the A91V and A136V in the signal receiver domain2529 and P116L at the first residue of the helix α516. Of these, polymorphisms A91V or A136V were detected in our nine overexpressed isolates (AB1, AB8, AB9, AB11, AB16, AB18, AB29, AB40 and AB44). Although the exact mechanisms need to be explained, such mutations in signal receiver domain of AdeR may change the interactions between the AdeS and AdeR and likely results in the adeABC overexpression. Furthermore, another mutation associated with multidrug resistance, L192R, was seen in AdeR of three adeB hyper-expressed isolates (AB22, AB35 and AB37). This mutation in the effector domain could alter protein stability30.
Several substitutions in AdeS were found to be responsible for adeABC overexpression: G30D located in the periplasmic loop31, the A94V and G103D alterations in the histidin kinase, adenylyl cyclase, methyl-accepting chemotaxis protein and phosphatase (HAMP) linker domain25, the G186V in the α-helix of the dimerization and histidine phosphotransfer (DHp) domain32 and T153M in the H box11. In the present study, while isolates AB16 and AB29 had polymorphisms A94V and G103D, respectively, there was coexistence of these mutations in five isolates AB3, AB9, AB13, AB35 and AB44. It is thought that such amino acid substitutions in the HAMP domain of AdeS protein disrupt transmembrane signal transduction process and have been suggested to be associated with constitutive phenotypes33. In addition, it is speculated that G186V mutation detected in four isolates AB1, AB11, AB18 and AB40 causes conformation changes of the AdeS DHp domain and then leading to activation of AdeS and stimulating the expression of the AdeABC efflux pump via interaction with AdeR. It is noteworthy that coexistence of such alterations in AdeR and AdeS TCS, regardless of whether is associated with other SNPs, may lead to efflux overexpression and affects drug susceptibility, synergistically.
In conclusion, our results suggest that the efflux-based system AdeABC is an important contributor to reduced susceptibility to antibiotics of choice for treatment, including ciprofloxacin and cefepime, in A. baumannii isolates. It is possible that the effective replacements together with the accumulation of SNPs in AdeR or AdeS may contribute to increased AdeABC expression and ciprofloxacin resistance, although the detailed effect of these variations on pump expression should be determined by rigorous experiments. Although adeB expression affects resistance to antimicrobials, gene expression alone is insufficient to develop multi-resistance, and so, multiple causes must be considered. These results may benefit to design active efflux pump inhibitors.
Acknowledgment
Authors acknowledge the laboratory staff at the Motahari Burns Hospital, Tehran, Iran for their cooperations.
Financial support & sponsorship: This study was financially supported by the Iran University of Medical Sciences, Tehran, Iran. (Grant no. 1067)
Conflicts of Interest: None.
References
- Global spread of drug-resistant Acinetobacter baumannii: Molecular epidemiology and management of antimicrobial resistance. Future Microbiol. 2011;6:407-22.
- [Google Scholar]
- Acinetobacter baumannii: An emerging opportunistic pathogen. Virulence. 2012;3:243-50.
- [Google Scholar]
- High prevalence of extensively drug-resistant and metallo beta-lactamase-producing clinical Acinetobacter baumannii in Iran. Microb Pathog. 2016;98:155-9.
- [Google Scholar]
- Acinetobacter baumannii clonal lineages I and II harboring different carbapenem-hydrolyzing-β-lactamase genes are widespread among hospitalized burn patients in Tehran. J Infect Public Health. 2015;8:533-42.
- [Google Scholar]
- Molecular epidemiology and characterization of multiple drug-resistant (MDR) clinical isolates of Acinetobacter baumannii. Int J Infect Dis. 2015;41:42-9.
- [Google Scholar]
- Mutation in the gyrA gene of quinolone-resistant clinical isolates of Acinetobacter baumannii. Antimicrob Agents Chemother. 1995;39:1201-3.
- [Google Scholar]
- Correlation of ciprofloxacin resistance with the AdeABC efflux system in Acinetobacter baumannii clinical isolates. Ann Lab Med. 2014;34:433-8.
- [Google Scholar]
- Mechanisms of resistance to ciprofloxacin, ampicillin/sulbactam and imipenem in Acinetobacter baumannii clinical isolates in Taiwan. Int J Antimicrob Agents. 2010;35:382-6.
- [Google Scholar]
- Association between mutations in gyrA and parC genes of Acinetobacter baumannii clinical isolates and ciprofloxacin resistance. Iran J Basic Med Sci. 2015;18:623-6.
- [Google Scholar]
- Resistance-nodulation-cell division-type efflux pump involved in aminoglycoside resistance in Acinetobacter baumannii strain BM4454. Antimicrob Agents Chemother. 2001;45:3375-80.
- [Google Scholar]
- Expression of the RND-type efflux pump AdeABC in Acinetobacter baumannii is regulated by the AdeRS two-component system. Antimicrob Agents Chemother. 2004;48:3298-304.
- [Google Scholar]
- Selection of topoisomerase mutations and overexpression of adeB mRNA transcripts during an outbreak of Acinetobacter baumannii. J Antimicrob Chemother. 2004;54:821-3.
- [Google Scholar]
- Characterization of the naturally occurring oxacillinase of Acinetobacter baumannii. Antimicrob Agents Chemother. 2005;49:4174-9.
- [Google Scholar]
- Clinical Laboratory Standards Institutes. Performance standards for antimicrobial susceptibility testing, 20th informational supplement. In: CLSI Document M100-S120. Wayne, PA: CLSI; 2013.
- [Google Scholar]
- Effect of efflux pump inhibitor carbonyl cyanide 3-chlorophenylhydrazone on the minimum inhibitory concentration of ciprofloxacin in Acinetobacter baumannii clinical isolates. Jundishapur J Microbiol. 2014;7:e8691.
- [Google Scholar]
- Tigecycline efflux as a mechanism for nonsusceptibility in Acinetobacter baumannii. Antimicrob Agents Chemother. 2007;51:2065-9.
- [Google Scholar]
- Distribution of the multidrug efflux pump genes, adeABC, adeDE and adeIJK, and class 1 integron genes in multiple-antimicrobial-resistant clinical isolates of Acinetobacter baumannii-Acinetobacter calcoaceticus complex. Int J Antimicrob Agents. 2009;33:27-32.
- [Google Scholar]
- Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: An international expert proposal for interim standard definitions for acquired resistance. Clin Microbiol Infect. 2012;18:268-81.
- [Google Scholar]
- Multidrug resistance among Acinetobacter baumannii isolates from Iran: Changes in antimicrobial susceptibility patterns and genotypic profile. Microb Drug Resist. 2014;20:632-40.
- [Google Scholar]
- Antimicrobial combinations against pan-resistant Acinetobacter baumannii isolates with different resistance mechanisms. PLoS One. 2016;11:e0151270.
- [Google Scholar]
- Antimicrobial resistance patterns and their encoding genes among Acinetobacter baumannii strains isolated from burned patients. Burns. 2012;38:1198-203.
- [Google Scholar]
- Molecular epidemiology and antimicrobial resistance determinants of multidrug-resistant Acinetobacter baumannii in five proximal hospitals in Taiwan. Jpn J Infect Dis. 2011;64:222-7.
- [Google Scholar]
- Multidrug resistant Acinetobacter baumannii – The role of AdeABC (RND family) efflux pump in resistance to antibiotics. Folia Histochem Cytobiol. 2008;46:257-67.
- [Google Scholar]
- Correlation of antimicrobial resistance with beta-lactamases, the OmpA-like porin, and efflux pumps in clinical isolates of Acinetobacter baumannii endemic to New York city. Antimicrob Agents Chemother. 2008;52:2999-3005.
- [Google Scholar]
- AdeABC-mediated efflux and tigecycline MICs for epidemic clones of Acinetobacter baumannii. J Antimicrob Chemother. 2010;65:1589-93.
- [Google Scholar]
- Modes and modulations of antibiotic resistance gene expression. Clin Microbiol Rev. 2007;20:79-114.
- [Google Scholar]
- Efflux-mediated antibiotic resistance in Acinetobacter spp. Antimicrob Agents Chemother. 2011;55:947-53.
- [Google Scholar]
- In vivo selection of a missense mutation in adeR and conversion of the novel blaOXA-164 gene into blaOXA-58 in carbapenem-resistant Acinetobacter baumannii isolates from a hospitalized patient. Antimicrob Agents Chemother. 2010;54:5021-7.
- [Google Scholar]
- AdeRS combination codes differentiate the response to efflux pump inhibitors in tigecycline-resistant isolates of extensively drug-resistant Acinetobacter baumannii. Eur J Clin Microbiol Infect Dis. 2014;33:2141-7.
- [Google Scholar]
- RND-type efflux pumps in multidrug-resistant clinical isolates of Acinetobacter baumannii: Major role for AdeABC overexpression and AdeRS mutations. Antimicrob Agents Chemother. 2013;57:2989-95.
- [Google Scholar]
- Screening and quantification of the expression of antibiotic resistance genes in Acinetobacter baumannii with a microarray. Antimicrob Agents Chemother. 2010;54:333-40.
- [Google Scholar]
- Single amino acid substitution Gly186Val in AdeS restores tigecycline susceptibility of Acinetobacter baumannii. J Antimicrob Chemother. 2016;71:1488-92.
- [Google Scholar]
- Mutational analysis of a conserved signal-transducing element: The HAMP linker of the Escherichia coli nitrate sensor NarX. J Bacteriol. 2003;185:89-97.
- [Google Scholar]






