Translate this page into:
Acute Q fever in individuals with acute febrile illness & exposure to farm animals: Clinical manifestations & diagnostic approaches
For correspondence: Dr Shilpshri V. Shinde, Department of Veterinary Public Health & Epidemiology, Nagpur Veterinary College, Maharashtra Animal & Fishery Sciences University, Nagpur, 440 006, Maharashtra, India e-mail: shilpi_shri5@rediffmail.com
-
Received: ,
Abstract
Background & objectives
Q fever is an important zoonotic disease affecting humans as well as animals. The objective of this study was to assess the burden of Q fever in individuals with acute febrile illness, particularly those in close contact with animals. Various diagnostic methods were also evaluated in addition to clinical examination analysis and associated risk factors.
Methods
Individuals presenting with acute febrile illness who had animal exposure were enrolled (n=92) in this study. Serum samples were tested using IgG and IgM phase 2 enzyme linked immunosorbent assay (ELISA) and immunofluorescence assay (IFA). The PCR targeting the com1 and IS1111 genes was performed on blood samples. PCR amplicons were sequenced and phylogenetically analysed. Demographic data, symptoms, and risk factors were collected through a structured questionnaire.
Results
Among individuals with acute febrile illness, 34.7 per cent (32 out of 92) were found to be infected with Coxiella burnetii. PCR exhibited the highest sensitivity among the diagnostic methods employed. The most common clinical manifestations included headache, chills, arthralgia, and fatigue. Individuals engaged in daily livestock-rearing activities were found to be at an increased risk of infection.
Interpretation & conclusions
Q fever is underdiagnosed due to its varied clinical presentations, diagnostic complexities, and lack of awareness. This study underscores the importance of regular screening for Q fever in individuals with acute febrile illness, particularly those with animal exposure. Early diagnosis and increased awareness among healthcare professionals are essential for the timely management and prevention of chronic complications associated with Q fever.
Keywords
Acute febrile illness
Q fever
animal exposure
ELISA
IFA
PCR
phylogeny
Q fever, caused by Coxiella burnetii, is an underreported and neglected zoonotic disease worldwide. Q fever is listed among the notifiable diseases by World Organization for Animal Health (WOAH) and is capable of infecting multiple animal species, including mammals, birds, and reptiles1. Humans are primarily infected through inhalation of aerosolized bacteria and also by ingestion. The disease can manifest in an acute or chronic form with an incubation period of 2-4 wk2. Around 60 per cent of the infected individuals are asymptomatic and, in the rest, it manifests as non-specific febrile illness in conjunction with hepatitis or pneumonia. Among these cases, 1-5 per cent develop as chronic infection characterized by endocarditis3. Veterinarians, farm, abattoir, and tannery workers, and those residing in or near livestock farm premises are at higher risk of infection4.
Indirect immunofluorescence assay (IFA) is considered as the gold standard to diagnose Q fever. A four-fold rise in the antibody titre in convalescent sera is confirmatory for Q fever5. As antibodies against C. burnetii are detectable only after 2-3 wks. of infection, it necessitates testing for both acute and convalescent-phase sera for serological diagnosis and hampers the early diagnosis of the disease6. PCR assay can detect the DNA of C. burnetii in a variety of samples, including blood, milk, and other clinical samples prior to seroconversion in both acute and chronic Q fever cases7. The combined application of IFA and PCR during the first two wk is believed to enhance the sensitivity of the diagnosis8.
Q fever is underdiagnosed because of limited clinical awareness, and the absence of definitive distinguishing features1. This study was undertaken to estimate the burden of C. burnetii infection in acute febrile illness cases at high risk of infection.
Material & Methods
Study design
This cross-sectional study was carried out over a period of 19 months from March 2020 to September 2021 in Vijayanagara district of Karnataka (2020) and Nagpur district of Maharashtra (2021). Samples were referred to the department of Veterinary Public Health, Nagpur Veterinary College, Nagpur from multiple tertiary care hospitals and primary health centres for screening of zoonotic diseases. Institutional ethical committee approval was obtained from Kamineni Academy of Medical Sciences and Research Centre, Hyderabad and Dr G.M. Taori Central India Institute of Medical Sciences, Nagpur. A written consent was obtained from all participating individuals before the start of the study.
Sampling
Individuals meeting the following inclusion criteria were enrolled in the study:
Clinical symptoms
Acute febrile illness with any other symptoms including headache, joint pain, chills, cough, myalgia, atypical pneumonia, and hepatitis with no definitive clinical diagnosis.
Epidemiological parameters
Individuals fulfilling any one of the following epidemiological inclusion criteria were included in the study:(i) having direct contact with livestock including activities such as milking, cleaning the shed, feeding, and watering of animals etc. (ii) presence of livestock in the household (iii) residing in the proximity to livestock farms (less than 1 km).
Exclusion criteria
Those individuals who had already received a confirmed diagnosis through laboratory tests for conditions such as COVID-19, dengue, typhoid, urinary tract infection, etc. were excluded. Additionally, those who exhibited clinical symptoms but did not meet the epidemiological criteria (i.e., had no contact with livestock) were also excluded from this study.
Only individuals fulfilling the clinical and epidemiological inclusion criteria were finally enrolled in the study. Demographic information, symptoms and risk factors related information of individuals were recorded (one to one interview or telephonic conversation) by use of a questionnaire (Supplementary material). A total of 92 individuals met the inclusion criteria. Blood samples were collected from the enrolled individuals and serum was separated. Serum samples were subjected to various serological tests.
PCR assay
DNA extraction from the blood samples was done using Himedia-HiPurA® Multi-Sample DNA Purification Kit as per manufacturer’s instructions. PCR targeting the com1 gene and nested PCR targeting the IS1111 gene were performed. Nested PCR was carried out using a set of external primers-Set I (Trans 1 and 2) and a set of internal primers Set II (Trans 3 and 4). The primer sequences and cycling conditions are presented in Supplementary Tables I and II, respectively. The amplified products were visualized in a 1.5 per cent agarose gel stained with ethidium bromide and observed under a gel documentation system (Bio-Rad laboratories, USA).
Serological assays
IgM and IgG enzyme linked immunosorbent assay (ELISA)
Coxiella burnetii (Q-Fever) Phase II NovaLisaTM IgM ELISA and Coxiella burnetii (Q-Fever) Phase II NovaLisaTM IgG ELISA kits were used to screen the serum samples. The tests were performed and interpreted as per manufacturer’s instructions. Each sample was diluted to 1:100 with a buffer, added to ELISA plate along with controls. The plate was incubated at 37°C for an hour, then washed. Conjugate was added to each well, followed by 30min incubation at 37°C. After washing, TMB substrate solution was added to all the wells and incubated in the dark for 15 min. Finally, stop solution was added. Samples ≥11 NTU value were considered positive. IgM phase II ELISA had diagnostic specificity of 95.95 per cent and a sensitivity of 95.74 per cent. In contrast, the IgG phase II ELISA had sensitivity and specificity of both at 100 per cent. As per manufacturers, the IgM Phase II ELISA kit may show cross-reactivity with dengue virus, cytomegalovirus, mycoplasma, and epstein-barr virus. Due to the possibility of false positives while using phase II IgM ELISA, the diagnosis of Q fever was further confirmed using additional diagnostic methods, such as PCR and IFA.
IgM and IgG IFA
The serum samples were screened using the Coxiella burnetii I+II IFA IgG/IgM/IgA kit (Vircell Microbiologists, Spain) as per the manufacturer’s protocol. Cut-off titre of ≥1/128 IgG Phase II antibodies and ≥1/24 IgM Phase II antibodies was considered for positive cases.
For detection of IgG antibodies reagents and slides were warmed to room temperature. Serum samples were diluted to 1:64 in PBS, with further two-fold dilutions made. Diluted samples and controls were added to slide wells and incubated at 37°C for 30 min. After washing and drying, anti-human IgG FITC conjugate solution was added and incubated again. Following another washing and drying, mounting medium was added and the slide was observed under a fluorescence microscope.
For IgM antibody detection, a 1:2 dilution was prepared and treated with anti-human IgM sorbent. Further two-fold dilutions were prepared up to 1:192. The diluted sample was added to wells, along with controls, and incubated at 37°C for 90 min. After washing and drying, anti-human IgM FITC conjugate solution was added and further processed as described above.
Diagnostic criteria
The diagnostic criteria given by CDC were used in the present study for diagnosis of acute Q fever9. Acute Q fever was diagnosed based on results of PCR, IFA and ELISA.
Blood and/or serum samples positive by PCR for either of the target gene (com1 and IS1111) regardless of IFA/ELISA results were considered as positive.
Serum samples showing a titre of ≥1:24 for IgM Phase II and ≥1:128 for IgG Phase II were considered positive for acute Q fever regardless of PCR results.
Serum samples with presence of either solitary IgG Phase II or IgM Phase II with negative PCR results were considered as inconclusive.
Sequencing and phylogenetic analysis
The PCR positive samples were sequenced (n=5; OM310954, OM310955, OM310958, OM310959, OM310960) in both the direction using com1F and com1R primers (Eurofins Genomics India, Bengaluru). The aligned sequences were submitted to NCBI database. Evolutionary analyses were conducted in MEGA11 software.
Statistical analysis
The statistical analysis was conducted in R statistical programme (Version 4.0.2). Univariate analysis was conducted using chi-square test for each of the risk factors. Odds ratios were calculated for the risk factors to determine the risk of being infected. Using IFAT as the gold standard, the sensitivity, specificity, and kappa values were calculated for ELISA and PCR. All statistical tests were performed at five per cent significance level and corresponding 95% confidence intervals. A P<0.05 was considered significant.
Results
Acute Q fever was diagnosed in 32 (34.7%) of the 92 participants using one of the tests (PCR, IFA and ELISA). Twenty four participants were positive for the com1 gene (PCR) and 26 were positive by the IS1111 gene (nested PCR). Overall, 28 (30.4%) participants showed expression of the specific genes. Seven were positive by PCR alone and did not show the presence of any anti-Coxiella spp antibodies. Individuals showing the presence of both IgM and IgG phase II antibodies were considered positive. Among the 92 samples screened using IFA, 27.1 per cent showed positivity for both IgM phase II and IgG phase II antibodies, thus, considered as cases of acute Q fever. Out of the 25 seropositive samples, four samples were negative by PCR. The samples were also screened by IgM phase II and IgG phase II ELISA which reported a seropositivity of 22.8 per cent and 4.3 per cent, respectively. All the ELISA positive samples were positive on IFA as well. Of the 92 samples screened using IFA and ELISA, three samples had inconclusive results due to the presence of solitary IgG phase II or IgM phase II and negative PCR results (Table I).
| Study area | Positive in PCR | Positive in IFA and/or ELISA | |||
|---|---|---|---|---|---|
| Antibodies against C. burnetii | No antibodies against C. burnetii |
Presence of both IgM Phase II and IgG Phase II antibodies (Positive in PCR) |
Presence of both IgM Phase II and IgG Phase II antibodies (Negative in PCR) | Presence of solitary IgG Phase II or IgM Phase II and negative PCR results | |
| Vijayanagara | 9 | 2 | 10 | 3 | 2 |
| Nagpur | 12 | 5 | 11 | 1 | 1 |
| Total | 21 | 7 | 21 | 4 | 3 |
| Grand total | 28 | 25 | --- | ||
| Diagnosed with acute Q fever | 34.7% (32/92) | ||||
PCR, polymerase chain reaction; IFA, immunofluorescene assay; ELISA, enzyme linked immunosorbent assay
Apart from pyrexia in Q fever positive participants, the most common symptoms were headache (56.4%), chills (43.1%), tiredness (37.9%), arthralgia (40.2%), and myalgia (35%). Rashes (6.66%), cough (3.33%) and lymphadenitis (9.5%) were also reported among the study participants (Table II). Symptoms such as diarrhoea, vomiting and night sweats were not reported by any of the participants.
| Symptoms | Patients reporting the symptoms (n) | positive cases n(%) |
|---|---|---|
| Myalgia | 80 | 28 (35) |
| Headache | 39 | 22 (56.4) |
| Chills | 58 | 25 (43.1) |
| Night sweating | 12 | 0 |
| Arthralgia | 67 | 27 (40.2) |
| Cough | 30 | 1 (3.33) |
| Rashes | 15 | 1 (6.66) |
| Fatigue | 79 | 30 (37.9) |
| Diarrhoea/stomach pain | 7 | 0 |
| Lymphadenitis | 21 | 2 (9.5) |
| Vomiting | 8 | 0 |
Higher percentage of the infection was observed in the age group of 36-45 yr and 46-55 yr. Higher positivity rate was observed in females (57.1%) as compared to males. Univariate analysis of risk factors is presented in Table III. The analysis revealed that individuals who had direct contact with animals and those who were engaged in practices such as cleaning of the shed, attending animals during parturition were at higher risk of developing C. burnetii infection. Practice of throwing placenta in lake and consumption of raw milk were also associated with the higher risk of infection. However, none of the risk factors identified were statistically significant.
| Parameters | Risk factor | Odds ratio | P value |
|---|---|---|---|
| Age group (yr) | 0-15 | - | 0.654 |
| 16-25 | |||
| 26-35 | |||
| 36-45 | |||
| 46-55 | |||
| 56-65 | |||
| >65 | |||
| Sex | Male | 0.599 | 0.5868 |
| Female | |||
| Consumption of raw milk | Yes | 2.058 | 0.502 |
| No | |||
| Practice slaughter of animal in house | Yes | 0.615 | 0.629 |
| No | |||
| Direct contact with animals | Yes | 2.38 | 0.514 |
| No | |||
| Animals in the household (but no direct involvement) | Yes | 2.15 | 0.22 |
| No | |||
| Cleaning the shed of animals | Yes | 2.67 | 0.56 |
| No | |||
| Assists animal in parturition | Yes | 1.4 | 1.00 |
| No | |||
| Disposal of placenta | In Lake | 4.5 | 0.1 |
| Burial | |||
| Ticks | Yes | 0.47 | 1.00 |
| No |
The PCR targeting the IS1111 and com1 genes revealed detection rates of 92.8 and 85.71 per cent, respectively. Nested PCR assay targeting the IS1111 gene was found to be more sensitive than conventional PCR targeting the same gene. PCR assays were found to be more effective in diagnosing acute cases of Q fever compared to the IFA and ELISA. The sensitivity, specificity, positive predictive and negative predictive values were 84, 100, 100and 94 per cent, respectively, when ELISA outcomes were compared with IFA. The kappa coefficient (0.88) indicated perfect agreement between ELISA and IFA. Similarly, when PCR assay was compared with IFA, the respective values were; 84 per cent sensitivity, 90 per cent specificity, 75 per cent positive predictive value, and 94 per cent negative predictive value . Substantial agreement was observed between PCR and IFA (Table IV). However, PCR had a better detection rate when compared to IFA in acute cases. Based on the calculated values, the true prevalence was found to be 27 per cent.
| Test | True prevalence (%) | Sensitivity (%) | Specificity (%) | PPV (%) | NPV (%) | Kappa coefficient |
|---|---|---|---|---|---|---|
| PCR | 27 | 84 | 90 | 75 | 94 | 0.70 |
| ELISA | 27 | 84 | 100 | 100 | 94 | 0.88 |
The statistical tests were performed at 5% significance level and corresponding 95% CI. PPV, positive predictive value; NPV, negative predictive value
The phylogenetic analysis revealed three distinct groups (Figure). Group I contained isolates from buffalo, cattle and ticks, group II had isolates from goat and hedgehogs, whereas, group III had isolates from mouse and cattle. All the sequences of the present study belonged to group I. The sequences exhibited a high sequence similarity (99-100%) with other isolates obtained from India. Furthermore, the sequences formed clusters with isolates collected from diverse hosts, and different origins.
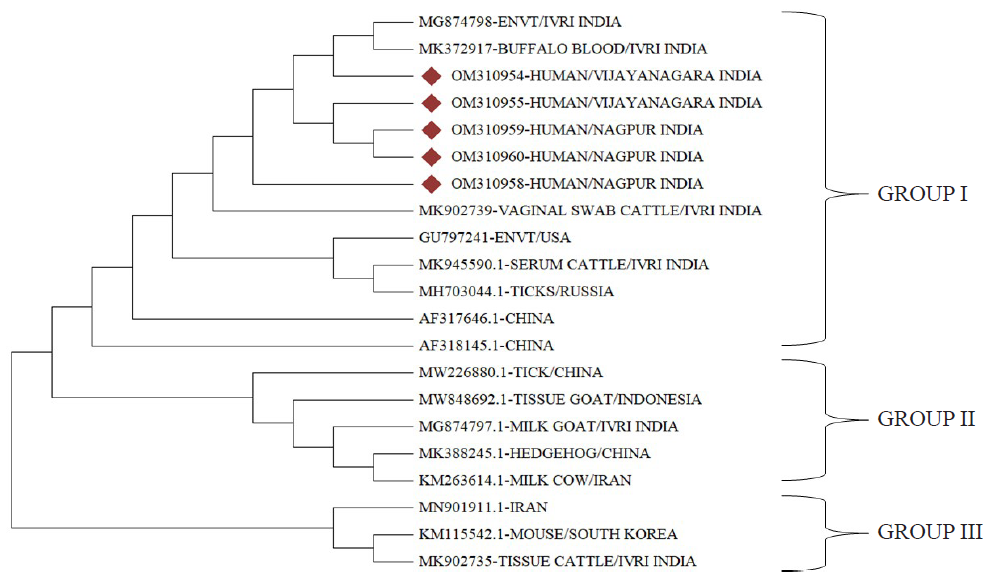
- Phylogenetic analysis using Neighbor joining method of sequenced samples (n=6) and com1 sequences from different parts of the world (n=17). The samples obtained in the study are indicated by red diamond symbol.
Discussion
Acute Q fever remains an underreported infection due to nonspecific symptoms in affected individuals, lack of awareness among physicians and diagnostic challenges10. The prevalence of acute Q fever in humans in India is not accurately known. Q fever is probably underestimated as 50-60 per cent of the individuals seroconvert without symptoms or report self-limiting mild febrile illness2. Reported seroprevalence in humans in India ranges from 46.7 to 89.47 per cent11,12. In India, there are limited studies that are focused on screening of individuals with febrile illness for acute Q fever2,13,14. This is in our knowledge one of the first systematic studies designed to estimate the burden of acute Q fever in high risk individuals with febrile illness in India.
Among the group of individuals with acute febrile illness and with history of regular animal contact, 34.7 per cent individuals were diagnosed with acute Q fever. A higher occurrence of Q fever among individuals who had frequent interactions with animals, particularly farm animals was reported. Individuals residing in close proximity to animal farms or rural areas are at an elevated risk of contracting the infection15,16. A high seropositivity (89.47%) for antibodies was reported among dairy farm workers from an organized dairy farm in Bareilly, India17. High seropositivity was attributed to prolonged contact with animals, large herd size, consumption of raw milk, and poor hygiene17. In the present study, majority of the participants reported having contact with animals indicating an increased risk of infection. Additionally, all the individuals resided within 1 km radius of animal habitats. C. burnetii can survive harsh environmental conditions. It has been reported that these bacteria can disperse over distances of up to 18 km from the source18, and a higher incidence of infection was observed among individuals residing within 5 km of the source19. These factors collectively might have contributed to high occurrence of acute Q fever in this study. A retrospective study in a tertiary care hospital in Pondicherry reported 9 out of 41 individuals with acute febrile illness for Q fever and among these five had contact with animals2. In Iran, out of 105 febrile individuals screened at a tertiary care centre using IFA, 37(35.2%) were diagnosed with acute Q fever and most had contact with livestock20. Acute Q fever is characterized by the sudden onset of high-grade fever accompanied by symptoms such as headache, myalgia, and fatigue and also typically involves a self-limiting flu-like illness21. In this study, the screened individuals reported headache, chills, fatigue, arthralgia, myalgia, and pyrexia as the commonly experienced symptoms. However, a subset of these also presented with additional symptoms such as rashes, lymphadenitis, and cough.
Higher occurrence of infection was reported among individuals aged 36-45 yr and 46-55 yr. Previous studies have reported a gradual increase in the occurrence of the disease with age, reaching a plateau around the age of 50 yr. Typically adults are actively engaged in livestock rearing activities, which exposes them to greater risks22. Females were found to have a higher proportion of infection as compared to males. Occupationally exposed females, involved in various farm tasks such as shed cleaning, manure collection, livestock care, milking, and animal feeding have been reported to be at an increased risk of contracting the disease23.
In the present study, several factors including contact with animals, cleaning of animal sheds, assisting animals during parturition, disposing of placenta in lakes, and consumption of raw milk were identified as significant risks for C. burnetii infection. These activities increase the chances of aerosol formation. Association of risk of exposure due to contact with the farm environment, rather than specific animal exposure, has been reported24.
Targeting the IS1111 gene resulted in a higher detection rate, which could be attributed to the presence of multiple copies of this gene in the bacterial genome. Detection of multicopy gene is advantageous as it ensures a higher initial concentration of the target sequence facilitating exponential amplification by PCR25. Nested PCR assay is reportedly 10 times more sensitive and specific than conventional PCR. In the present study, higher detection rate was observed in the trans-nested PCR when compared to conventional PCRs26. In diagnosing acute cases of Q fever, the PCR assay proved to be more sensitive than IFA and ELISA. PCR, as a diagnostic tool for acute Q fever, is highly specific and useful during the initial two wk of infection27. Use of PCR in screening individuals suspected of acute Q fever, even when antibodies against the pathogen are not present has been reported2. The use of PCR and IFA for screening suspected acute Q fever samples is recommended, citing the unreliability of results from commercial ELISA kits28. IFA confirmed acute Q fever with a four-fold rise in serum titre. However, in the absence of convalescent sera, the detection of both IgM phase II and IgG phase II can serve as diagnostic criteria for acute Q fever2,9.
Phylogenetic analysis revealed that the sequences of isolates from various geographic regions and multiple host species clustered together. This observation suggests that despite the diversity of origins, these sequences share common evolutionary relationships. The com1 gene, is highly conserved and homologous; a previous study conducted in India also reported similar sequence clustering29. These findings strongly suggest a high level of conservation of the com1 gene within the bacterial genome.
The major limitation of this study was the inability to perform follow up of the screened individuals. Hence, a detailed study involving paired sera samples could not be performed. Detailed history of the participants could not be collected as the samples were sent to the department for screening and details were often collected through telephonic conversation with the patient. Since the study was conducted mostly during the period of COVID 19 pandemic, it was difficult to interact with the participants to collect history and other details; thus, only a few participants met the enrolment criteria.
C. burnetii infections are often underdiagnosed and underreported, but these have been increasingly identified in recent years25. In this study, samples were collected from individuals with acute febrile illness having contact with farm animals, leading to a higher detection rate of acute Q fever. Despite the limited sample size, the study provided evidence of the occurrence of Q fever in India, highlighting its public health importance. The results suggested that persons closely associated with livestock were at a higher risk of infection. PCR is recommended for early detection, with IFA for confirmation. Raising awareness among high-risk groups and physicians is vital for early disease detection. High-risk individuals should hence be informed about transmission routes and farm biosecurity measures to prevent disease spread.
Financial support & sponsorship
This study was supported by Indian Council of Medical Research, New Delhi (IRIS ID No- 2020-5169 RFCNo.P-54/ECD/Adhoc/7/2021-22dated 23.02.2022).
Conflicts of Interest
None.
Use of Artificial Intelligence (AI)-Assisted Technology for manuscript preparation
The authors confirm that there was no use of AI-assisted technology for assisting in the writing of the manuscript and no images were manipulated using AI.
References
- Current perspectives on the occurrence of Q fever: Highlighting the need for systematic surveillance for a neglected zoonotic disease in Indian subcontinent. Environ Microbiol Rep. 2021;13:138-58.
- [Google Scholar]
- Detection of acute Q fever human cases by indirect immunofluorescence & real-time polymerase chain reaction in a tertiary care hospital in Puducherry. Indian J Med Res. 2018;148:449-452.
- [Google Scholar]
- Current approaches for the detection of Coxiella burnetii infection in humans and animals. J Microbiol Methods. 2020;179:106087.
- [Google Scholar]
- Seroprevalence of Q fever in the United States, 2003–2004. Am J Trop Med Hyg. 2009;81:691-4.
- [Google Scholar]
- High prevalence of Coxiella burnetii infection in humans and livestock in Assiut, Egypt: A serological and molecular survey. Vet World. 2020;13:2578-86.
- [Google Scholar]
- Diagnostic usefulness of molecular detection of Coxiella burnetii from blood of patients with suspected acute Q fever. Medicine (Baltimore). 2019;98:e15724.
- [Google Scholar]
- Diagnosis of acute Q fever by detection of Coxiella burnetii DNA using real-time PCR, employing a commercial genesig easy kit. J Clin Diagn Res. 2017;11:DC10-3.
- [Google Scholar]
- Presentation and diagnosis of acute Q fever in Portugal — A case series. IDCases. 2017;7:34-7.
- [Google Scholar]
- Diagnosis and management of Q fever — United States, 2013: Recommendations from CDC and the Q fever working group. MMWR Recomm Rep. 2013;62:1-29.
- [Google Scholar]
- Isolation of Coxiella burnetii in patients with nonspecific febrile illness in South Korea. BMC Infect Dis. 2020;20:421.
- [Google Scholar]
- Apparent prevalence and risk factors of coxiellosis (Q fever) among dairy herds in India. PLOS One. 2020;15:e0239260.
- [Google Scholar]
- Apparent prevalence and risk factors associated with occurrence of coxiella burnetii infection in goats and humans in Chhattisgarh and Odisha, India. Comp Immunol Microbiol Infect Dis. 2018;60:46-51.
- [Google Scholar]
- Bartonella quintana and Coxiella burnetii as causes of endocarditis, India. Emerg Infect Dis. 2008;14:1168-9.
- [Google Scholar]
- First genetic evidence of Coxiella burnetii in cases presenting with acute febrile illness, India. J Med Microbiol. 2017;66:388-90.
- [Google Scholar]
- Prevalence of coxiella burnetii infection in humans occupationally exposed to animals in Poland. Vector-Borne Zoonotic Dis. 2015;15:261-7.
- [Google Scholar]
- The sero-epidemiology of Coxiella burnetii in humans and cattle, Western Kenya: Evidence from a cross-sectional study. PLoS Negl Trop Dis. 2016;10:e0005032.
- [Google Scholar]
- Seroprevalence and molecular detection of coxiellosis among cattle and their human contacts in an organized dairy farm. J Infect Public Health. 2019;12:190-4.
- [Google Scholar]
- Surveillance of Coxiella burnetii shedding in three naturally infected dairy goat herds after vaccination, focusing on bulk tank milk and dust swabs. Vet Sci. 2022;9:102.
- [Google Scholar]
- Airborne geographical dispersal of Q fever from livestock holdings to human communities: A systematic review and critical appraisal of evidence. BMC Infect Dis. 2018;18:218.
- [Google Scholar]
- Acute Q fever among febrile patients in Zahedan, Southeastern Iran. Turk J Med Sci. 2014;44:99-103.
- [Google Scholar]
- Q Fever: An old but still a poorly understood disease. Interdiscip Perspect Infect Dis. 2012;2012:e131932.
- [Google Scholar]
- Febrile patients admitted to remote hospitals in Northeastern Kenya: Seroprevalence, risk factors and a clinical prediction tool for Q-Fever. BMC Infect Dis. 2016;16:244.
- [Google Scholar]
- Cross-sectional study for determining the prevalence of Q fever in small ruminants and humans at El minya governorate, Egypt. BMC Res Notes. 2017;10:538.
- [Google Scholar]
- Coxiella burnetii seroprevalence and risk factors in cattle farmers and farm residents in three northeastern provinces and inner mongolia autonomous region, China. BioMed Res Int. 2016;2016:e7059196.
- [Google Scholar]
- Molecular detection and typing of Coxiella burnetii. Available from: https://www.rivm.nl/bibliotheek/rapporten/330291002.pdf, accessed on July 5, 2023.
- Evaluation of PCR and nested PCR assays currently used for detection of coxiella burnetii in Japan. Southeast Asian J Trop Med Public Health. 2004;35:852-5.
- [Google Scholar]
- Epidemiological, clinical and laboratory features of acute Q fever in a cohort of hospitalized patients in a regional hospital, Israel, 2012-2018. PLoS Negl Trop Dis. 2021;15:e0009573.
- [Google Scholar]
- Serological evidence of spotted fever group rickettsiosis in and around Puducherry, south India—A three years study. J Vector Borne Dis. 2018;55:144-50.
- [Google Scholar]
- Coxiella burnetii in cattle and their human contacts in a gaushala (cattle shelter) from India and its partial com1 gene sequence-based phylogenetic analysis. Anim Biotechnol. 2021;33:1449-58.
- [Google Scholar]






